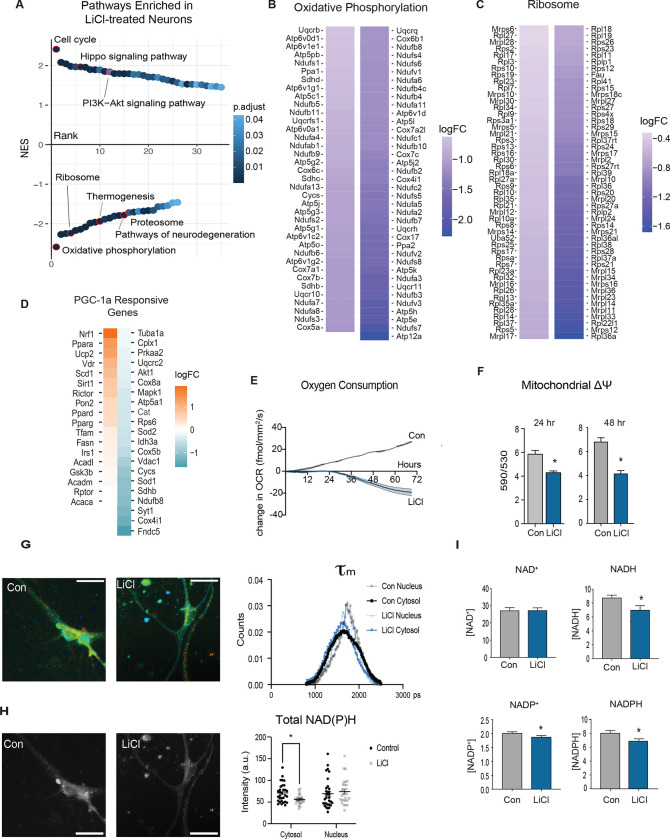
Figure 5: Neuronal metabolic response to GSK3b inhibition with LiCl.
A) Rank order plot of enriched pathways detected by GSEA of RNA-sequencing of neurons treated for 24 hours with 15mM LiCl (n=4). B-C) Heatmap of the significantly changing genes in the oxidative phosphorylation (B) and ribosome (C) pathways enriched in LiCl-treated neurons. D) Heatmap of transcripts detected in the LiCl-treated neuron RNA-sequencing data set known to be responsive to changes in PGC-1a expression ranked by logFC. E) Oxygen consumption of primary neurons treated with LiCl or a control media change measured by RESIPHER live-cell oxygen consumption monitor for 72 hours (n=6). F) Mitochondrial membrane potential assessed by JC-1 assay in primary neurons treated for 24 and 48 hours with 15mM LiCl. G) Representative images (left)and distributions (right) of NAD(P)H mean fluorescence lifetime of control or LiCl-treated primary neurons. H) Representative images (left) and quantitation (right) of NAD(P)H fluorescence intensity of control or LiCl-treated primary neurons. I) NAD, NADH, NADP, and NADPH biochemical assays. Asterisks indicate p-value <0.05 by student’s t-test (F, H, and I) and multiple Mann-Whitney tests (E).