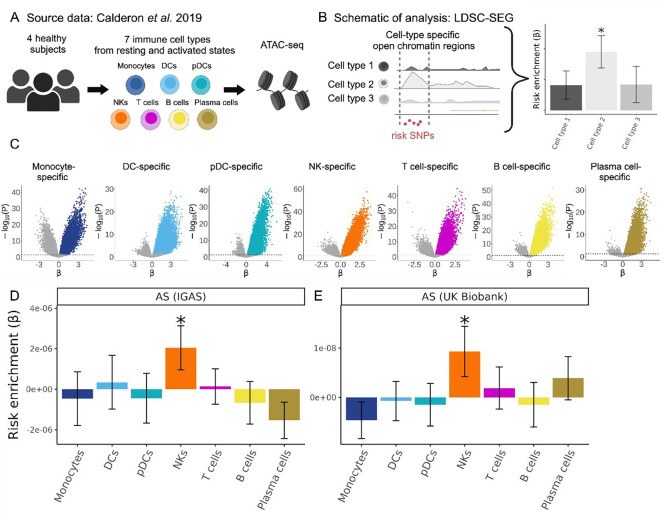
Figure 1. Human NK cell-specific open chromatin regions are enriched in AS genetic risk.
(A) Cartoon depicting the Calderon et al. study design. Peripheral blood cells from 4 healthy subjects were sorted into immune cell populations that we grouped in silico into seven cell types (see Methods). Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) was performed with and without prior in vitro activation. (B) Graphical representation of LDSC-SEG analysis: identification of cell type-specific annotations (in our case open chromatin regions), followed by the integration with GWAS summary statistics to obtain a risk enrichment coefficient β and P-value. (C) Volcano plots showing results of differential accessibility analyses for each cell type compared to the other cell types. Colored dots indicate open chromatin peaks in the top decile of the t-statistic for each cell type, which were used for LDSC-SEG analysis. (D-E) Bar graphs displaying the AS genetic risk enrichment coefficient β and block jackknife standard error for cell type-specific open chromatin accounting for control peaks and baseline annotations. Summary statistics from the International Genetics of Ankylosing Spondylitis Consortium (IGAS) (D) and UK Biobank (E) GWAS were used. * indicates P < 0.05.