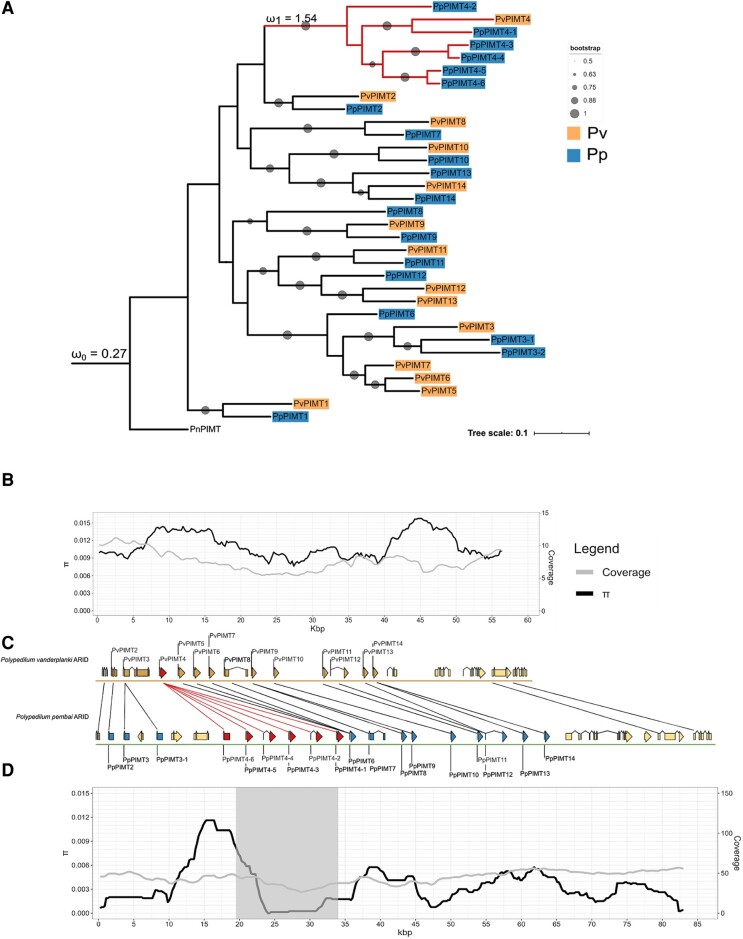
Fig. 2.
Comparative analysis of the PIMT ARId from two midges. (A) ML tree with bootstrap testing (100 replicates). Pv, P. vanderplanki; Pp, P. pembai, Pn, P. nubifer. Red color shows the clade under positive selection; blue color shows P. pembai PIMTs; orange color shows P. vanderplanki PIMTs. (B) Nucleotide diversity along P. vanderplanki PIMT ARId; (C) Comparison of gene order in the two ARId. Black lines represent homologous genes; (D) Nucleotide diversity along the P. pembai PIMT ARId. Gray color shows the zone of selective sweep found by Pool-hmm. Coverage curve shown in (A, D) is based on mapping of Tashan nabai population genomic data of P. vanderplanki and Chikopa population genomic data of P. pembai. ω1 stands for the foreground ratio, and ω0, for the background ratio.