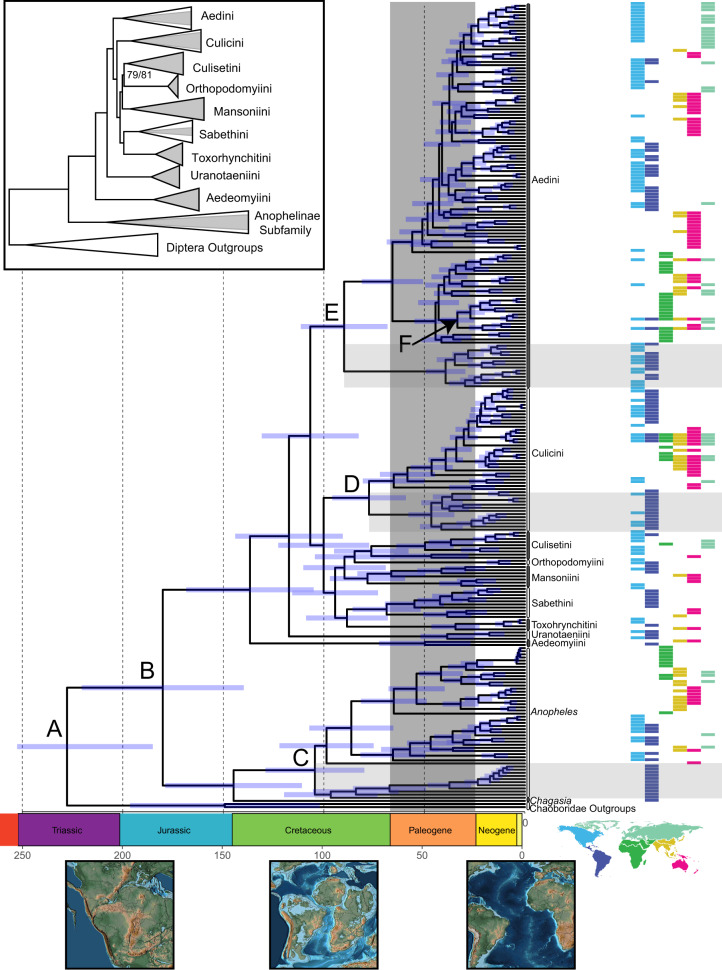
Fig. 1. Phylogenetic relationships among major lineages of mosquitoes (Culicidae), as inferred by maximum likelihood and dated using a fossil-calibrated relaxed clock analysis.
The analysis is based on the analysis of amino acid sequences of 709 genes (525,000 aligned sites) from 256 species and calibrated at seven time points30,70,94–98. Two Chaoborid outgroups were used to root the phylogeny. Horizontal gray boxes indicate major clades confined to the Americas, whose divergence times correlate with the dates of major geological events. Bars on nodes are 95% HPDs. Paleogeographic reconstructions are from PALEOMAP99, and colored bars correspond to the biogeographic region of the species listed, as indicated in the world map below. In the inset, tribes have been collapsed for easier viewing, and the proportion of genera sampled are indicated by gray triangles. Support values are SH-like aLRT and Ultrafast Bootstrap values and are only shown if branch support was below 99. A The common ancestor of the Chaoboridae and the Culicidae. B The common ancestor of all extant Culicidae. C The common ancestor of all Anopheles in our analysis. D The common ancestor of all Culex in our analysis. E The common ancestor of the tribe Aedini. F The common ancestor of all Aedes (Stegomyia) sampled in our analysis. Lettered annotations are discussed in greater detail in the text.