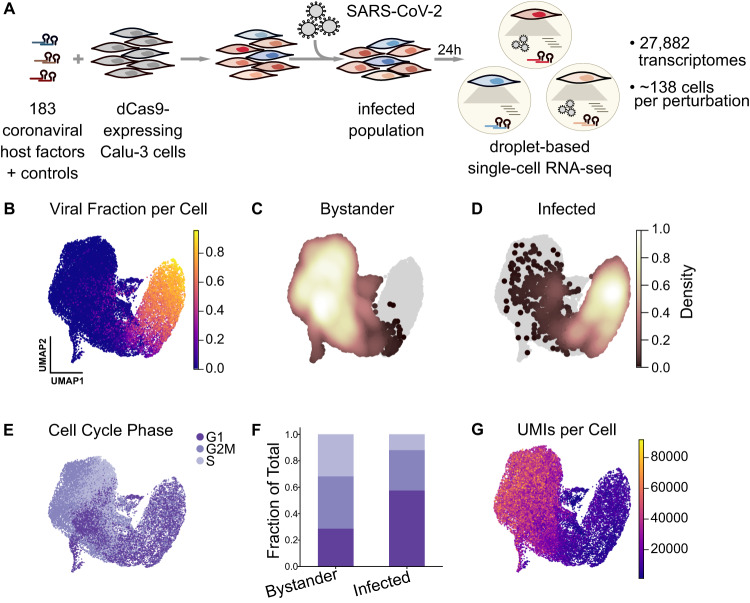
Fig. 1. Perturb-seq for single-cell transcriptional analysis and functional validation of SARS-CoV-2 host factors.
A Experimental design for the Perturb-seq experiment in Calu-3 cells engineered to express CRISPRi machinery. We perturbed 183 different host factors (individually or in combination) using a lentivirally-delivered library, infected the cells with SARS-CoV-2 for 24 hours, and performed droplet-based single-cell RNA sequencing, reading out host and viral transcripts as well as the sgRNA, indicating the perturbed host factor. B Single-cell transcriptomes were projected into UMAP space and colored by viral RNA fraction per cell. C, D Density of cells identified as either uninfected/bystander (C) or infected (D) by our classifier, overlaid onto all cells in gray. E Cells color-coded by their cell cycle phase. F Fraction of bystander and infected cells assigned to each cell cycle phase. G Cells color-coded by the number of detected UMIs per cell.