Figure 6.
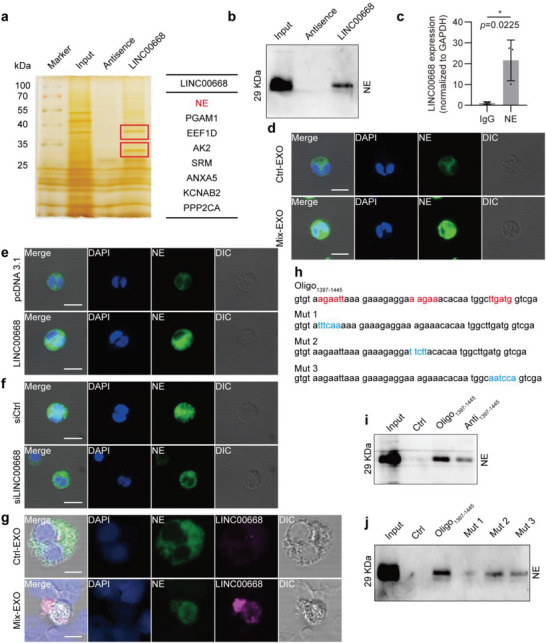
LINC00668 promotes the translocation of NE from the cytoplasmic granules to the nucleus and induces NETs release. a) The proteins interacting with LINC00668 in HL‐60 cells identified by biotinylated LINC00668 pull‐down followed by mass spectrometry. b) The proteins associated with LINC00668 in HL‐60 cells were pulled down by biotinylated LINC00668 or its antisence sequence (Antisence), and the precipitates were analyzed by Western blotting using anti‐NE antibody. c) qRT‐PCR detection of LINC00668 in the RNA‐protein immunoprecipitates pulled down by IgG or anti‐NE antibody in LINC00668‐overexpressing HL‐60 cells. d) Neutrophils were incubated with the exosomes isolated from Ctrl (Ctrl‐EXO) or Mix‐treated (Mix‐EXO) Caco‐2 cells, and immunofluorescence staining detected the subcellular localization of NE. Green and blue staining indicates NE and the nuclei, respectively. Differential Interference Contrast (DIC) shows 3D stereoscopic projection of cells. Scale bars represent 50 µm. e) Neutrophils were transfected with pcDNA3.1 or LINC00668‐overexpressing plasmids, and immunofluorescence staining detected the subcellular localization of NE. Green and blue staining indicates NE and the nuclei, respectively. Scale bars represent 50 µm. f) Caco‐2 cells were transfected with siCtrl or siLINC00668 and stimulated with Mix for 24 h. Exosomes were isolated from the culture supernatant and used to treat neutrophils, and NE subcellular localization was detected using immunofluorescence staining. Scale bar represents 50 µm. g) Immunofluorescence staining of NE and LINC00668 fluorescence in situ hybridization (FISH) on neutrophils treated with the exosomes isolated from Ctrl (Ctrl‐EXO) or Mix‐treated (Mix‐EXO) Caco‐2 cells. Green, magenta, and blue staining indicate NE, LINC00668, and the nuclei, respectively. Scale bars represent 50 µm. h) The sequence of 1397–1445 nucleotide positions in the LINC00668 sequence (Oligo1397‐1445) and the 3 mutants (Mut 1, Mut 2, and Mut 3) for the binding sites 1, 2, and 3 in Oligo1397‐1445 sequence. i) The lysates of HL‐60 cells were incubated with biotinylated Oligo1397‐1445, the antisense sequence of Oligo1397‐1445 (Anti1397‐1445) or Ctrl, and Western blot analysis detected NE level in the Oligo‐protein immunoprecipitates. j) HL‐60 cell lysates were incubated with biotinylated Oligo1397‐1445 or the mutants for the binding sites 1, 2, and 3 (Mut 1, Mut 2, and Mut 3), and Western blot analysis detected NE level in the Oligo‐protein immunoprecipitates. Data are represented as mean ± SEM. *p<0.05, p‐value was determined by unpaired two‐tailed Student's t‐test.
