Abstract
Object
This study aims to investigate the changes in gut microbiota and metabolism of patients with chronic kidney disease (CKD) stage 1–2, as well as the potential impact of hyperuricemia (HUA) on these factors in CKD stage 1–2 patients.
Methods
In this study, fecal samples were collected from CKD stage 1–2 without HUA patients (CKD-N group), CKD stage 1–2 with HUA patients (CKD-H group), and healthy people controls (HCs group). The samples were then subjected to the microbiome (16S rRNA gene sequencing) and metabolome (liquid chromatography-tandem mass spectrometry) analyses. The multi-omics datasets were analyzed individually and integrated for combined analysis using various bioinformatics approaches.
Results
Gut microbial dysbiosis was found in CKD-N and CKD-H patients. At the phylum level, compared to HCs group, Bacteroidetes decreased but Proteobacteria increased in CKD-H group significantly. Fusobacteria in CKD-N group was significantly lower than HCs group. At genus level, [Eubacterium]_ventriosum_group, Fusobacterium, Agathobacter, Parabacteroides, and Roseburia significantly changed in CKD groups. [Ruminococcus]_gnavus_group was significantly lower in CKD-H group than CKD-N group. Moreover, the fecal metabolome of CKD-N and CKD-H altered significantly. d-glutamine and d-glutamate metabolism, arginine and proline metabolism, histidine metabolism, and lysine biosynthesis were down-regulated in the CKD-N group. Phenylalanine metabolism, arginine and proline metabolism, purine metabolism, and beta-alanine metabolism were up-regulated in the CKD-H group. There was a significant difference between the two CKD groups in phenylalanine metabolism. The abundance change of [Ruminococcus]_gnavus_group, [Eubacterium]_ventriosum_group, UCG-002, Alistipes, and Bifidobacterium had a close correlation with differential metabolites.
Conclusion
The gut microbiota and metabolic status undergo significant changes in CKD patients compared to healthy people. Additionally, HUA has been found to impact the gut microbiota of CKD patients, as well as their metabolism. The close association between gut microbiota and metabolites suggests that the former plays a crucial role in metabolism.
Keywords: Early stage chronic kidney disease, Hyperuricemia, Gut microbiota, Metabolome
1. Introduction
Chronic kidney disease (CKD) is a significant public health concern worldwide, with 8–16% prevalence among adults [1]. The kidney play a crucial role as the primary excretory organ for uric acid [2], responsible for eliminating approximately 70% of the uric acid produced by human body. Hence, when renal function loss, the prevalence of hyperuricemia(HUA)increases [3]. Meanwhile, HUA is considered an independent factor causing CKD [4].
Recent studies have indicated that CKD patients may experience changes in their gut microbiota. Specifically, an analysis of early-stage CKD patients revealed a significant increase in the abundance of Actinobacteria and Proteobacteria [5]. Besides, studies have shown that CKD patients have a decreased abundance of Firmicutes, Trichococcaceae, Ruminaceae, and Faecobacteriaceae, while the abundance of Bacteroidetes, Proteobacteria, Clostridium, and Enterobacteriaceae increased significantly. Interestingly, the “gut-kidney axis” theory suggests that dysbiosis of gut microbiota can lead to symptoms of kidney inflammation [6]. There appears to be a reciprocal relationship between CKD and gut microbiota imbalances, with CKD leading to microbiota imbalances and the imbalances contributing to CKD progression. HUA may also influence gut microbiota, as Ren [7] found that HUA caused an increased of Firmicutes and a decrease of Bacteroidetes and Proteobacteria in HUA mice. However, there is little research on how both CKD and HUA impact gut microbiota composition.
CKD and HUA may cause the change of metabolic status. It was reported that tryptophan decreased but its metabolite kynurenine increased in CKD patients [8]. Previous studies have demonstrated that HUA affects 138 metabolites based on untargeted serum metabolomics [9]. Among these metabolites, serine, glutamate, and glutamine were down-regulated, while glycine, hydroxyproline, and alanine were up-regulated in hyperuricemia nephropathy mice [10]. However, there is a dearth of research on the collective impact of CKD and HUA on metabolism. However, there are few studies on the metabolic impact of CKD and HUA.
Therefore, In order to address the aforementioned issues, we collected fecal samples from patients in CKD stages 1–2 with HUA, as well as from healthy individuals. Our aim was to investigate the alterations in gut microbiota and metabolism in early CKD with HUA, using 16S rRNA gene sequencing and LC-MS/MS.
2. Results
2.1. Patients with CKD 1–2 and HUA display altered microbiota composition compared to healthy subjects
All participants, including patients and volunteers, were of Han Chinese ethnicity and resided in the Shanghai area, with similar dietary habits. The clinical variables of the three groups were shown in Table 1.
Table 1.
Basic characteristics and biochemical indexes of participants in three groups. *P < 0.001, compared to HCs group; #P < 0.001, compared to CKD-H group eGFR: estimate glomerular filtration rate; SCr: serum creatinine; SUA: serum uric acid; CysC: cystatin C, TG: triglycerides; TC: total cholesterol, HDL: high density lipoprotein; LDL: low density lipoprotein, Hb: hemoglobin; ACR: urinary albumin to creatinine ratio.
| HC(n = 15) | CKD-N(n = 15) | CKD-H(n = 15) | |
|---|---|---|---|
| Gender(Male/Famale) | 6/9 | 10/5 | 15/0 |
| Age(years) | 30.6 ± 5.7 | 45.53 ± 8.82 | 42.73 ± 13.39 |
| eGFR(ml/min.1.73m2) | 104.01 ± 9.14 | 88.08 ± 25.48* | 79.60 ± 12.12* |
| Scr(umol/l) | 71 ± 12.72 | 85.4 ± 23.5* | 102.33 ± 13.54* |
| SUA(umol/l) | 325.8 ± 52.51 | 325.73 ± 54.2# | 530.2 ± 54.02* |
| CysC(umol/l) | 0.59 ± 0.07 | 1.103 ± 0.26* | 1.19 ± 0.42* |
| TC(umol/l) | 4.36 ± 0.59 | 5.28 ± 2.92 | 4.04 ± 1.56 |
| TG(umol/l) | 1.37 ± 0.29 | 2.05 ± 1.19 | 2.36 ± 2.12 |
| HLD(umol/l) | 1.49 ± 0.26 | 1.169 ± 0.38 | 0.86/0.31 |
| LD(umol/l) | 2.42 ± 0.68 | 3.70 ± 2.78 | 2.54 ± 0.99 |
| Hb(g/l) | 137.93 ± 11.04 | 141 ± 15.61 | 146.2 ± 11.09 |
| ACR(mg/g) | 17.80 ± 6.50 | 150 ± 68.2* | 74.71 ± 18.73* |
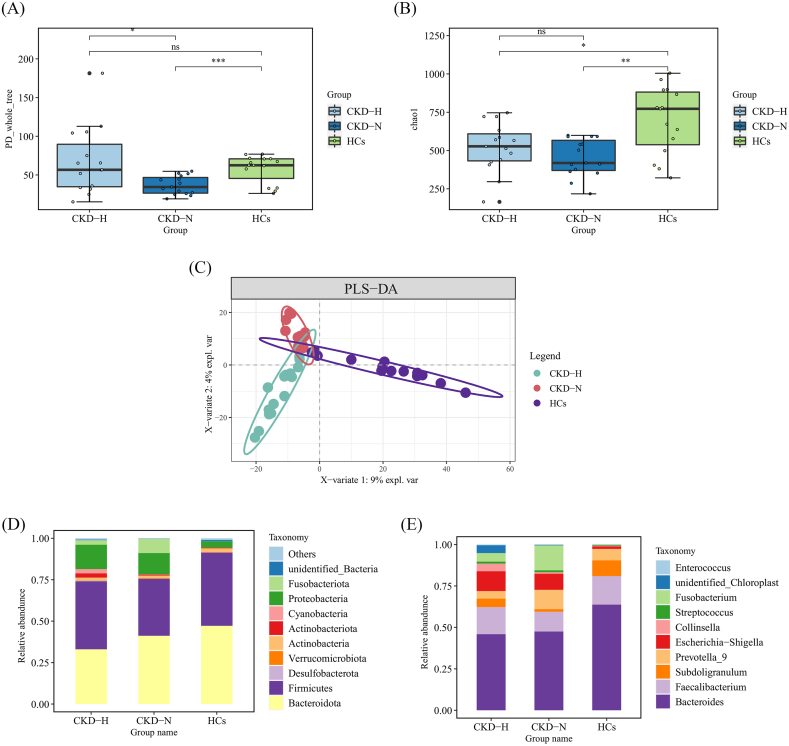
In view of the influence of CKD and HUA on the gut microbiota, we explored the structure and differences in the gut microbiota of patients. A total of 4055 OTUs were obtained after assigned and annotated. The α-diversity of the gut microbiota was estimated by PD_whole_tree index and chao1 index. The former reflected the community richness and evenness,and the latter reflected the community richness. The PD_whole_tree indexes of both CKD-N and CKD-H groups were significantly lower than those of HCs. Additionally, CKD-H group had significantly higher indexes than CKD-N group (HCs: 57.7 ± 18.0, CKD-N: 36.7 ± 11.9, CKD-H:67.8 ± 43.4; CKD-N vs HCs: p = 0.000997; CKD-H vs HCs: p = 0.0128) (Fig. 1A). Similarly, chao1 of CKD-N and CKD-H group were significantly lower than those of HCs (HCs: 696 ± 220.0, CKD-N: 453.1 ± 122.2, CKD-H: 520.1 ± 160.3; CKD-N vs HCs: p = 0.003; CKD-H vs HCs: p = 0.026). (Fig. 1B). β-diversity among the three groups was evaluated using PLS-DA, which showed a clear difference in the distribution of the groups (Fig. 1C). PCoA and NMDS were also employed to analyze β-diversity (Supplementary Fig. 1).
Fig. 1.
Comparison of the bacterial structure between the HCs, CKD-N and CKD-H group. (A, B)The box plots of (A) PD_whole_tree and (B) chao1 index showed the α-diversity of three groups. (C) PLS-DA was used to analyzed the β-diversity of three groups. (D, E) Bar diagrams presented the relative abundance of top 10 (D) phyla and (E) genera.
Moreover, we specially focused on the top 10 most abundant phyla (Fig. 1 D) and general (Fig. 1E). Bacteroidetes and Firmicutes were the dominant phyla in each group. Compared to HCs (47.1%), the abundance of Bacteroidota decreased both in CKD-N (41.1%) and CKD-H groups (33.0%). Compared to HCs (44.2%), while the abundance of Firmicutes decreased slightly in CKD-N (34.4%), it increased in CKD-H group (41.1%) under the influence of HUA. Meanwhile, compared to HCs group, the relative abundance of Proteobactertia, Fusobacteriota, Actinobacteriota and Cyanobacteria rose obviously both in CKD-N and CKD-H groups. Furthermore, the relative abundance of Fusobacteriota decreased in CKD-H group compared to CKD-N group while Actinobacteriota increased in CKD-H group (HCs: 3.8%, 0.08%, 0.4%, 0.04%; CKD-N:12.8%, 8.6%, 0.6%, 0.2%; CKD-H: 14.7%, 0.3%, 2.5%, 0.2%). At genus level, Bacteroides was the dominant genus in all participants which decreased slightly in CKD-N group and CKD-H group (HCs: 31.3%; CKD-N:27.1%; CKD-H: 24.6%). Escherichia-Shigella and Fusobacterium increased in CKD-N and CKD-H groups, and the latter was more abundant in CKD-N group (HCs: 0.73%, 0.08%; CKD-N:5.6%, 8.6%; CKD-H: 6.4%, 2.8%). There were obvious increase of Collinsella in CKD-H group and decrease of Subdoligranulum in CKD-N group comparing to the other two groups.
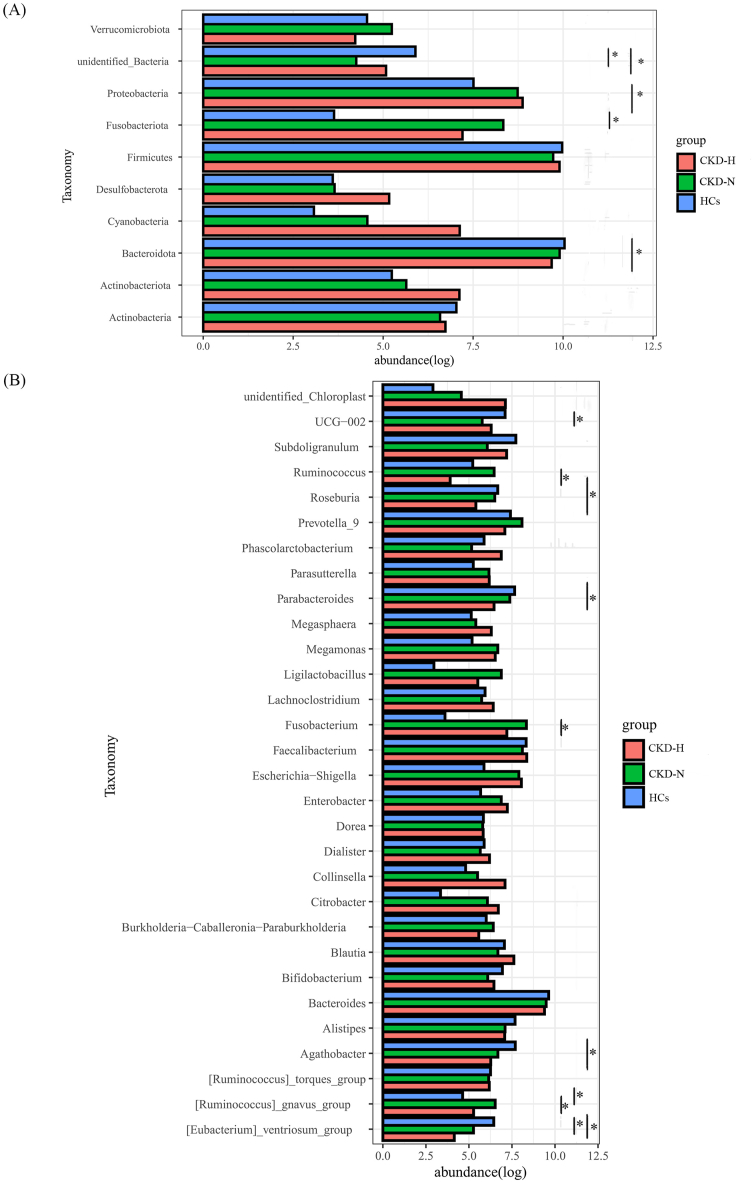
In order to compare the differences among these three groups, t-test between two groups was performed three times and the results were corrected by Bonferroni method. At the phylum level (Fig. 2A), it were observed a slight decrease of Bacteroidota was observed in CKD-N group compared to HCs group and a significant decrease in CKD-H group. The abundance of Proteobactertia in CKD-H groups and Fusobacteriota in CKD-N were significantly higher than HCs, but there was no significant difference in these taxa between CKD-N and CKD-H groups. At genus level (Fig. 2B), 30 genera were compared. Compared to HCs group, [Eubacterium]_ventriosum_group decreased significantly both in CKD-N and CKD-H groups. Agathobacter, Parabacteroides, and Roseburia decreased significantly in CKD-H group. In CKD-N group, the abundance of Fusobacterium increased but UCG-002 decreased significantly. What's more, the relative abundance of [Ruminococcus]_gnavus_group significantly higher in CKD-N group than HCs, but under the influence of HUA the relative abundance of [Ruminococcus]_gnavus_group decreased in CKD-H group.
Fig. 2.
The relative abundance of top 10 phyla and top 30 genera and the comparison results between two groups. T-test was performed to check the significant difference between two groups. *: p < 0.05 (A, B) The histogram plots presented the abundance and comparison results of (A) top 10 phyla and (B) top 30 genera.
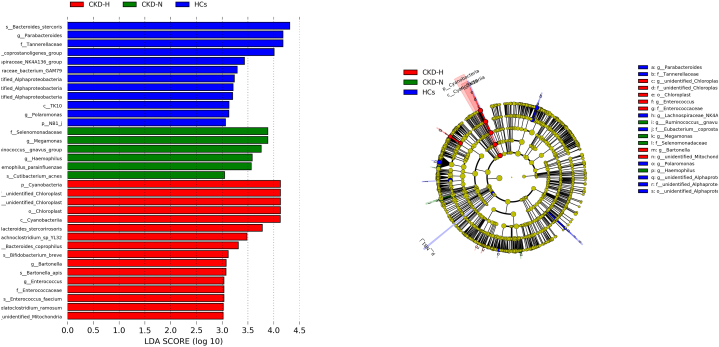
The results of the linear discriminant analysis (LDA) effect size (LEfSe) showed that g_Parabacteroides, s Bacteroides_stercoris, f_Tannerellaceae were the biomarkers in HCs group. Meanwhile, g_Megamonas and g_Ruminococcus gnavus group were the biomarkers in CKD-N group. And p_Cyanobacteria and g_Enterococcus were the biomarkers in CKD-H group, as illustrated in Fig. 3.
Fig. 3.
The results of LEfSe analysis.
2.2. Evaluation of fecal metabolites reveals alterations in the gut metabolome in CKD 1–2 patients with HUA
The fecal samples were analyzed using the UHPLC-QE-MS analytical technique, and after data cleaning, 13,229 peaks were identified for further analysis. The OPLS-DA analysis showed a distinct separation in the distribution of sample metabolites between each two groups (Fig. 4A (a)-(c)). Metabolites which VIP >1 in OPLS-DA and p value < 0.05 in student's t-test were regarded as differential expressed metabolites (DEMs).
Fig. 4.
The status of metabolism in three groups. All the results were got by comparing two groups. (a) comparison was between HCs and CKD-N groups; (b) comparison was between HCs and CKD-H groups; (c) comparison was between CKD-N and CKD-H groups. Positive value means the pathways or metabolites are up-regulated in the latter group, vice versa. (A) OPLS-DA analysis between two groups. (B) DA score was chose as the index to describe the relative abundance of metabolism pathways between two groups. (C) Bubble plots presented the differential metabolism pathways between two groups. There were significant differences in labeled pathways.
The heatmap of 201 DEMs indicated significant differences in fecal metabolic patterns between CKD-N and HCs groups. DEMs between HCs and CKD-N showed distinct clustering in each group. Similarly, the heatmap of 155 DEMs revealed significant differences in fecal metabolic patterns between CKD-H and HCs groups. 45 DEMs between CKD-N and CKD-H groups were identified in differential metabolic pathways (Supplementary Fig. 3).
These differential metabolites were assigned to different metabolic pathways according to KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway database. Differential abundance score was used to describe the change situation of metabolism pathways. The metabolic pathways of 201 DEMs between CKD-N and HCs groups were analyzed. These pathways included amino acid metabolism(including tryptophan metabolismand histidine metabolism), carbohydrate metabolism (including glyoxylate and dicarboxylate metabolism and carbon metabolism), digestive system(including vitamin digestion and absorption and protein digestion and absorption), lipid metabolism (including linoleic acid metabolism and arachidonic acid metabolism), metabolism of cofactors and vitamins (including d-glutamine and D−glutamate metabolismand beta−alanine metabolism) and others(Fig. 4B (a)). Metabolic pathways of 155 DEMs between CKD-H and HCs included amino acid metabolism(including arginine and proline metabolism and phenylalanine metabolism), digestive system(bile secretion), lipid metabolism(including primary bile acid biosynthesis and glycerophospholipid metabolism), metabolism of cofactors and vitamins (porphyrin and chlorophyll metabolism), nucleotide metabolism (including purine metabolism and pyrimidine metabolism, neuroactive ligand−receptor interaction) (Fig. 4B (b)). However, 45 DEMs between CKD-H and CKD-H were failed to be assigned to specific metabolic pathways (Fig. 4B (c)).
As depicted in Fig. 4C (a), the majority of pathways exhibited down-regulation in the CKD-N group in comparison to the HCs. Notably, the down-regulation of d-glutamine and d-glutamate metabolism, arginine and proline metabolism, histidine metabolism and lysine biosynthesis displayed a significant statistical difference. In CKD-H group, there was a significant increase in phenylalanine and beta-alanine metabolism, while arginine and proline metabolism and purine metabolism were significantly decreased (Fig. 4C (b)). The only statistically significant difference between the CKD-N and CKD-H groups was the up-regulation of phenylalanine metabolism in the CKD-H group. (Fig. 4C (c)).
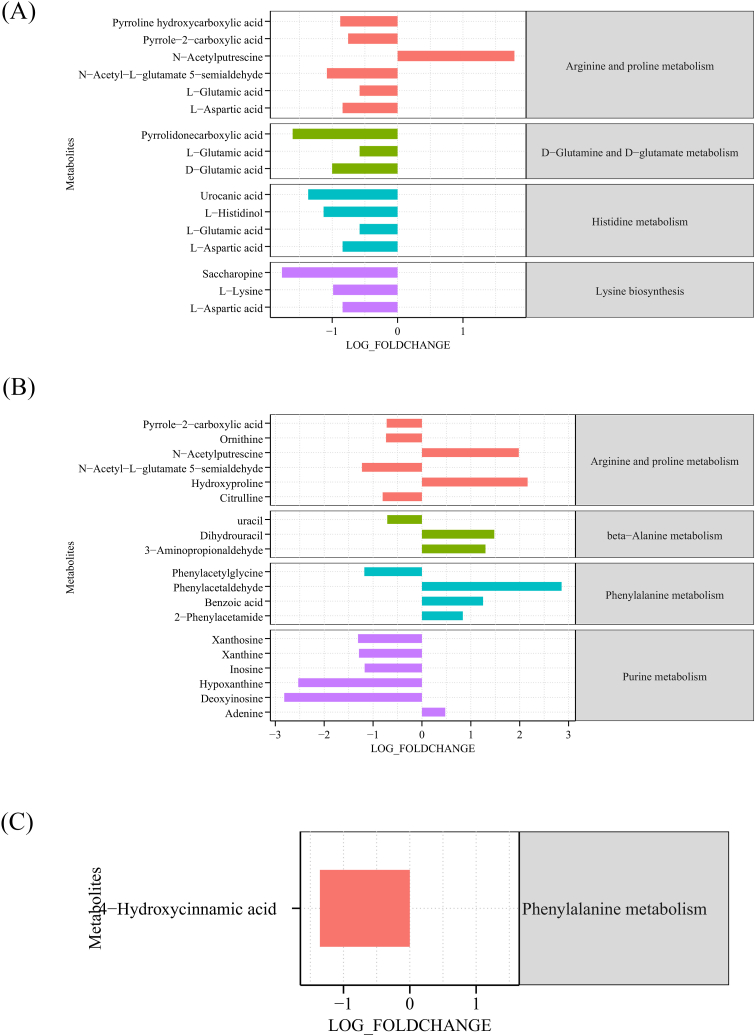
The specific metabolites change in the pathways was shown in Fig. 5. Compared to HCs group, the majority of metabolites in mentioned pathways in CKD-N group down-regulated, with the exception of N-acetylputrescine in arginine and proline metabolism (Fig. 5A). Similarly, in the CKD-H group, metabolites belonging to purine metabolism, except adenine, were down-regulated when compared to the HCs group (Fig. 5B). However, compared to CKD-N groups, 4-hydroxycinnamic acid was down-regulated (Fig. 5C).
Fig. 5.
The differential metabolites between (A) HCs and CKD-N groups, (B) CKD-N and CKD-H groups, (C) CKD-N and CKD-H groups in the metabolic pathways. The log (fold change) was used to presented the abundance difference and positive value means the pathways or metabolites are up-regulated in the latter group, vice versa.
2.3. Potential correlation of differential microbial abundances and metabolic profiling alterations was primarily analyzed
Based on the above gut microbiota and metabolomics data, correlation analysis was performed to identify microbe-associated metabolites in each group. Only the microbiota detected in more than half of groups was selected. The results were shown in Supplementary Fig. 4. The results of correlation of metabolites belong to metabolic pathways with significant differences and the top 30 abundant genera were show in Fig. 5. Spearman algorithm was performed to calculate the correlation coefficient and p value. Heatmaps were plotted by R (V 4.2.2).
In Fig. 6A, the correlation of top 30 genera and metabolites in HCs groups was showed. [Eubacterium]_ventriosum_group and [Ruminococcus]_gnavus_group were found to be the most strongly associated with these metabolites, and interestingly, their correlation with the metabolites was almost opposite to each other [Ruminococcus]_gnavus_group was significantly negatively correlated with d-glutamic acid, l-glutamic acid and pyrrolidonecarboxylic acid, the differential metabolites in d-glutamine and d-glutamate metabolism. Besides, pyrroline hydroxycarboxylic acid, N-acetyl-l-glutamate 5-semialdehyde and saccharopine were significantly negatively correlated with [Ruminococcus]_gnavus_group [Eubacterium]_ventriosum_group was positively correlated with inosine, deoxyinosine, and hypoxanthine which all took part in purine metabolism.
Fig. 6.
Correlation of 30 most abundant genera and metabolites in metabolism pathways with significant difference. * mean the correlation had statistic significance. (A) The correlation of gut microbiota and metabolites in HCs group. (B) The correlation of gut microbiota and metabolites in CKD-N group. (C) The correlation of gut microbiota and metabolites in CKD-H group.
In Fig. 6B, UCG-002 showed close correlation with metabolites. UCG-002 was positively correlated with xanthine and uracil, but negatively correlated with phenylacetaldehyde, benzoic acid, and 2-phenylacetamide, which took part in phenylalanine metabolism. The biomarker Megamonas was negatively correlated with urocanic acid, xanthosine, and L-histidinol. Ruminococcus gnavus group was found positively correlated with inosine, adenine, and dihydrouracil.
As for CKD-H group, Alistipes and Bifidobacterium were most deserving mentioned. The former was also negatively correlated with phenylacetaldehyde and benzoic acid. The latter was negatively correlated with inosine, xanthosine, xanthine, and hypoxanthine, indicating the influence of Bifidobacterium on purine metabolism(Fig. 6C).
3. Discussion
Akin to an anaerobic bioreactor, the colorectum harbors an enormous diversity of microbiota, which are capable of producing an extraordinarily wide range of small molecules (i.e., short chain fatty acids) that influence many vital pathways associated with energy homeostasis, nutritional intake, and immune balance [11]. The results of this study indicate that compared to healthy people, both CKD and CKD with HUA patients have obvious changes in gut microbiota and fecal metabolites. Meanwhile, the differential metabolites between HCs and CKD patients have close relation to gut microbiota.
Previous studies reported that gut microbiota dysbiosis has been observed in individuals with both CKD and HUA. Ye found that when compared to healthy people, the abundance of Firmicutes, Lachnospiraceae, Ruminobacteriaceae and Faeobacteriaceae decreased in CKD patients, but Bacteroidetes, Proteobacteria, Clostridium, and Enterobacteriaceae significantly increased [12]. It was also found the gut microbiota of mice with HUA exhibited significant alterations in both species and abundance when compared to that of normal mice. The abundance of Bacteroidetes and Proteobacteria decreased, while Firmicutes and Actinobacteria increased [13]. The findings of this study provide evidence of a notable imbalance in gut microbiota between healthy individuals and CKD patients. At phylum level, the abundance of Bacteroidota decreased and Proteobacteria increased significantly in CKD-H patients. According to Yang [14], the decrease of Bacteroidota was also found in asymptomatic HUA patients. The increase of Proteobacteria was also reported by Hu [15], which was consistent of our study. Xi [16]reported the presence of numerous pathogens in gout has been associated with an increase in Proteobacteria. LEfse analysis indicated Cyanobacteria was one of biomark of CKD-H group. Cyanobacteria may produce Microcystin-LR, a kidney toxin that could potentially explain the link between Cyanobacteria and CKD [17]. At the genus level, compared to healthy people, some pathogens increased in CKD patients. The increase of [Ruminococcus]_gnavus_group has statistic significance which was thought associated with inflammatory bowel disease in CKD patients. [Ruminococcus]_gnavus_group demonstrated a strong ability to differentiate CKD [18] and HUA [19]. Meanwhile, some SCFA producers, such as [Eubacterium]_ventriosum_group, Parabacteroides, and Roseburia decreased in CKD patients, indicating the risk of intestinal barrier dysfunction. The increase of Fusobacterium in CKD patients was reported and the abundance of Fusobacterium was positively correlated with serum uric acid [20]. Interestingly, the abundance of [Ruminococcus]_gnavus_group decreased significantly in CKD-H group, compared to CKD-N group. The function of [Ruminococcus]_gnavus_group was controversial [21]. The effects of it on HUA need further research.
Previous studies have demonstrated gut microbiota and their metabolites play significant role in the pathogenesis of HUA and gout [22], it suggested that gut microbiota may influence host health status through metabolites. The results showed that diverse pathways mainly include d-glutamine and d-glutamate metabolism, arginine and proline metabolism, lysine biosynthesis, histidine metabolism and alanine, aspartate and glutamate metabolism, among which arginine and proline metabolites emerged as the most significantly altered, and N-acetylputrescine was the only increased metabolite involved in arginine and proline metabolites. Interestingly, N-acetylputrescine was positively associated with renal fibrosis [23]. Therefore, we speculate an increase in N-acetylputrescine could trigger and worsen kidney disease. Besides, histidine metabolism reduced in CKD patients. There is evidence to suggest that histidine possessed both antioxidant and anti-inflammatory properties, and that a decrease in histidine levels may increase the risk of various metabolic syndromes [24]. It was worth noting that l-glutamate is involved in multiple metabolic pathways as shown in Fig. 3D, which was reported played important role in protection epithelial cells from oxidative and inflammatory stress [25]. The decrease of l-glutamate levels in CKD patients implied the risk of intestinal epithelial cell dysfunction. Other literature pointed out that dietary l-lysine prevented adenine-induced arterial calcification in uremic rats [26]. The decreased levels of l-lysine in CKD patients may contribute to their increased risk for cardiovascular disease [27].
Phenylalanine metabolism, beta-alanine metabolism, arginine and proline metabolism, and purine metabolism were different between CKD-H and HCs groups. Phenylalanine metabolism was proposed as a key pathway associated with HUA to induce CKD. 4-hydroxycinnamic acid, which is involved in phenylalanine metabolism, was down-regulated in CKD-H groupcompared to CKD-N group. 4-Hydroxycinnamic acid has anti-inflammatory and immunomodulatory properties [28]. The reduction in 4-hydroxycinnamic acid mean that patients with both CKD and HUA were at a higher risk of developing inflammation [29].
Part of gut microbiota was found closely associated with metabolism. [Eubacterium]_ventriosum_group and [Ruminococcus]_gnavus_group were both correlated with the metabolites participating d-glutamine and d-glutamate metabolism and purine metabolism. The increment of [Ruminococcus]_gnavus_group lead to oxidative stress and intestinal barrier damage, which may influence metabolism [30]. Alistipes was negatively correlated with phenylacetaldehyde and benzoic acid, and it was reported that Alistipes was positively correlated with hydrocinnamic acid, which participated in phenylalanine metabolism [31], showing close relation with phenylalanine metabolism. Bifidobacterium was negatively correlated with purine metabolism, which was consist of the report in obese people [32].
There are several limitations of this study. Firstly, the sample size of each group may not be large enough to avoid coincidences. Secondly, age and gender were not matched across the different groups, which may have influenced systemic metabolism. Futhermore, the correlation between differential metabolites and gut microbiota was draw from data analysis perspective, which was need more experiments to validate.
4. Conclusion
In conclusion, the gut microbiota of individuals with CKD are quite different from healthy people and the presence of HUA influences the gut microbiota of CKD patients. The abundance of [Ruminococcus]_gnavus_group significantly decreased under the influence of HUA, compared to CKD with normal serum uric acids level patients. As for metabolism, arginine and proline metabolism is different from HCs and CKD patients. The difference of phenylalanine metabolism between CKD-N and CKD-H groups is significant. The change of [Eubacterium]_ventriosum_group, [Ruminococcus]_gnavus_group, Alistipes, and Bifidobacterium may influence metabolism in CKD patients. Thus, the change of gut microbiota induced by CKD and HUA may influence the health status of patients.
5. Materials and methods
5.1. Patients and samples
Patients with stage 1–2 CKD and HUA (CKD-H,n = 15) and patients with stage 1–2 CKD without HUA (CKD-N, n = 15) were recruited from Nephrology Department of Minhang Hospital Affiliated to Fudan University (Shanghai, China) between April 2021 and October 2021. A control group of healthy individuals (HCs, n = 15) were recruited from the medical examination center of the same hospital. All participants were Han Chinese living in Shanghai with similar eating habits. CKD patients received standardized diets during hospitalization. Healthy volunteers had normal bowel habits and no history of CKD or HUA. All participants were informed of the study objectives and methods.
Patient inclusion criteria: All subjects aged between 18 and 75 years. According to KDIGO criteria CKD 1–2 stage was defined as the patients whose eGFR ≥60 ml/min/1.73 m2 and have albuminuria, urine sediment abnormalities, or structural abnormalities on imaging. Albuminuria is defined as a urine albumin-to-creatinine ratio (ACR) ≥30 mg/g or a urine protein-to-creatinine ratio (PCR) ≥150 mg/g. Urine sediment abnormalities refer to the presence of red blood cells, white blood cells, or casts in the urine. Structural abnormalities on imaging refer to abnormalities detected on ultrasound, CT, or MRI scans of the kidneys. The patients in CKD-N group were CKD 1–2 stage patients and the levels of serum uric acid in male patients were less than 420umol/l in male patients, less than 360umol/l in female patients. The patients in CKD-H group were CKD 1–2 stage patients and the levels of serum uric acid in male patients were higher than 420umol/l in male patients, higher than 360umol/l in female patients.
Exclusion criteria: Subjects complicated with gastroenteritis or mother major digestive system diseases tumor in the past 3 months were excluded. Subjects had diabetes or aotoimmune diseases were excluded. Besides, subjects had received antibiotic, prebiotic, or probiotic in a month prior to recruiting were excluded.
The studies involving human participants were reviewed and approved by Institutional ethics board of Fudan University Minhang Hospital. This study was approved by our local ethics committee (ethics approval number 2021-056-01K).
5.2. Sample collecting
Qualified stool samples were self-collected in sterile frozen tubes by subjects and were transported immediately to the laboratory where they were stored at −80 °C for further testing. Routine clinical parameter tests were performed in the clinical laboratories of Minhang Hospital Affiliated to Fudan University.
5.3. 16S rRNA gene sequencing
Frozen stool samples were shipped on dry ice and stored at −80 °C until DNA extraction, which was performed using CTAB/SDS method. The DNA concentration and purity were assessed using a 1% agarose gel, and the DNA was subsequently diluted to a final concentration of 1 μg/μL using sterile water16S rRNA genes of distinct regions(16S V4) were amplified using specific primer (16S V4: 515F–806R) with the barcode. All PCR reactions were carried out with 15 μL of Phusion® High-Fidelity PCR Master Mix (New England Biolabs); 0.2 μM of forward and reverse primers, and about 10 ng template DNA. Thermal cycling consisted of initial denaturation at 98 °C for 1 min, followed by 30 cycles of denaturation at 98 °C for 10 s, annealing at 50 °C for 30 s, and elongation at 72 °C for 30 s. A final elongation step at 72 °C for 5 min was performed to ensure complete extension of the amplified product.
Sequence libraries were generated using the TruSeq® DNA PCR-Free Sample Prep Kit (Illumina, USA) according to the manufacturer's recommendations, and index codes were added. Libraries were assessed for quality using a Qubit® 2.0 Fluorometer (Thermo Scientific) and an Agilent Bioanalyzer 2100 system. Finally, the library was sequenced on the Illumina NovaSeq platform, resulting in 250 bp paired-end reads. Paired-end reads were assigned probes and merged using FLASH (VI.2.7). Perform quality filtering on raw labels to obtain high-quality clean labels on QIIME (V1.9.1). Compare these tags with the Silva database Chimeric sequences were deleted using the UCHIME algorithm. Sequences with more than 97% similarity were classified as identical operational taxonomic units (OTUs) using UPARSE (V 7.0.1001). Using the Silva database, each representative sequence was annotated with taxonomic information based on the Mothur algorithm. Alpha diversity and beta diversity were calculated using QIIME software (V 1.9.1). Before cluster analysis, Bray-Curtis distance-based non-metric multidimensional scaling analysis (NMDS) was performed using the vegan package in R software.
5.4. Untargeted fecal metabolomics analysis
Untargeted metabolomics analysis of stool samples by liquid chromatography-tandem mass spectrometry (LC-MS/MS). A UPLC-BEH-Amide column (2.1 mm × 100 mm, 1.7 μm) was connected to a Q Exactive HFX mass spectrometer (Orbitrap MS, Thermo) using a UHPLC system (Vanquish, Thermo Fisher Scientific). Results of separate analysis of positive and negative ions were merged.
We used Metabolanalyst (https://www.metaboanalyst.ca) to perform data cleaning, statistical analysis, and path enrichment analysis. In order to reduce the impact of the detection system error on the results and make the results better highlight the biological significance, we have made a series of preparations and managements for the original data, including: Outlier filtering: Individual peaks are filtered to remove noise. Based on the relative standard deviation (RSD), that is, the coefficient of variation (CV), the deviation is filtered. Missing value filtering: Filter on a single Peak. Only the peak area data with no more than 50% null values in a single group or no more than 50% null values in all groups is kept. Missing value recording: Missing values in the original data are recorded by filling in the half of the minimum value.
Data normalization: Normalization was performed by internal standard (IS).
In more than 50% of the samples, the peak intensities were removed from the peak intensity matrix. The remaining missing values are replaced by one-fifth of the smallest positive value for each variable. If the relative standard deviation of the deviation is greater than 25%, it is filtered and normalized by the mean. Orthogonal projection latent structure discriminant analysis (OPLS-DA) algorithm, fold change (FC) and t-test were used to identify metabolites that differed significantly between groups. Permutation tests (100 permutations) were used to validate the OPLS-DA model. Differentially expressed metabolites (DEM) were identified by stringent criteria, ie variable importance (VIP) value in p rejection >1, log2 (fc) > |2|, p < 0.05. Metaboanalyst refined the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and identified it as a potential target with a threshold of p < 0.05.
5.5. Statistics analysis
All normally distributed data are presented as mean ± standard deviation and statistically testing by GraphPad Prism software (V9.1.2). To compare two groups, Wilcoxon rank-sum test and unpaired Student's t-tests were utilized and the results were corrected by Bonferroni method. LEfse analysis was performed using LEfse tool (http://huttenhower.sph.harvard.edu/galaxy/) with an alpha value < 0.05 and LDA score >3.0. Figures were visualized using the R (V4.2.2) software.
Funding statement
This study was supported by the Traditional Chinese Medicine Research Project of Shanghai Municipal Health Commission (2020JP011); Natural Science Foundation Project of Shanghai Municipal Science and Technology Commission (21dz1200200);Shanghai Minhang District Characteristic Specialty Construction Project (No.2020MWTZB07) by grants from Key Research and Development Program of Anhui Province (No. 202004j07020049).
Consent for publication
All authors approved the manuscript for publication.
Author contribution statement
Ping Liu and Jianli Yang: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper; Contributed reagents, materials, analysis tools or data. Yifan Zhu and Yuyan Tang: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data. Yu Chen: Conceived and designed the experiments; Wrote the paper. Xudong Xu and Haidong He: Conceived and designed the experiments; Analyzed and interpreted the data.
Data availability statement
Data associated with this study has been deposited at SRA of NCBI under the accession number RJNA879265.
Ethics statement
The studies involving human participants were reviewed and approved by Institutional ethics board of Fudan University Minhang Hospital (ethics approval number 2021-056-01K). The patients/participants provided their written informed consent to participate in this study.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e20328.
Contributor Information
Xudong Xu, Email: xxdmzx@sina.com.
Haidong He, Email: chinahhd@sina.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Hu Q., Wu K., Pan W., Zeng Y., Hu K., Chen D., Huang X., Zhang Q. Intestinal flora alterations in patients with early chronic kidney disease: a case-control study among the Han population in southwestern China. J. Int. Med. Res. 2020;48 doi: 10.1177/0300060520926033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson R.J., Bakris G.L., Borghi C., Chonchol M.B., Feldman D., Lanaspa M.A., Merriman T.R., Moe O.W., Mount D.B., Sanchez Lozada L.G., Stahl E., Weiner D.E., Chertow G.M. Hyperuricemia, acute and chronic kidney disease, hypertension, and cardiovascular disease: report of a scientific workshop organized by the national kidney foundation. Am. J. Kidney Dis. 2018;71:851–865. doi: 10.1053/j.ajkd.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Russo E., Viazzi F., Pontremoli R., Barbagallo C.M., Bombelli M., Casiglia E., Cicero A.F.G., Cirillo M., Cirillo P., Desideri G., D'Elia L., Ferri C., Galletti F., Gesualdo L., Giannattasio C., Iaccarino G., Leoncini G., Mallamaci F., Maloberti A., Masi S., Mengozzi A., Mazza A., Muiesan M.L., Nazzaro P., Palatini P., Parati G., Rattazzi M., Rivasi G., Salvetti M., Tikhonoff V., Tocci G., Ungar A., Verdecchia P., Virdis A., Volpe M., Grassi G., Borghi C. U. Working Group on, and H. Cardiovascular Risk of the Italian Society of, Association of uric acid with kidney function and albuminuria: the uric acid right for heart health (URRAH) project. J. Nephrol. 2021 doi: 10.1007/s40620-021-00985-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.K. Tada, T. Maeda, K. Takahashi, K. Ito, T. Yasuno, S. Funakoshi, A. Satoh, M. Kawazoe, C. Yoshimura, S. Mukoubara, K. Masutani, H. Arima, and H. Nakashima, Association between serum uric acid and new onset and progression of chronic kidney disease in a Japanese general population: iki epidemiological study of atherosclerosis and chronic kidney disease.Clin. Exp. Nephrol. 9.. [DOI] [PubMed]
- 5.Felizardo R.J., Castoldi A., Andrade-Oliveira V., Camara N.O. The microbiota and chronic kidney diseases: a double-edged sword. Clin Transl Immunology. 2016;5:e86. doi: 10.1038/cti.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrios C., Beaumont M., Pallister T., Villar J., Goodrich J.K., Clark A., Pascual J., Ley R.E., Spector T.D., Bell J.T., Menni C. Gut-microbiota-metabolite axis in rarly renal function decline. PLoS One. 2015;10 doi: 10.1371/journal.pone.0134311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ren Q., Cheng L., Guo F., Tao S., Zhang C., Ma L., Fu P. Fisetin improves hyperuricemia-induced chronic kidney disease via regulating gut microbiota-mediated tryptophan metabolism and aryl hydrocarbon receptor activation. J. Agric. Food Chem. 2021;69:10932–10942. doi: 10.1021/acs.jafc.1c03449. [DOI] [PubMed] [Google Scholar]
- 8.Zakrocka I., Zaluska W. Kynurenine pathway in kidney diseases. Pharmacol. Rep. 2022;74:27–39. doi: 10.1007/s43440-021-00329-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin N., Jiang Y., Shi W., Wang L., Kong L., Wang C., Guo Y., Zhang J., Ma Q. High-Throughput untargeted serum metabolomics analysis of hyperuricemia patients by UPLC-Q-TOF/MS. Evid. base Compl. Alternative Med. 2021;2021:1–15. doi: 10.1155/2021/5524772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan L., Han P., Ma S., Peng R., Wang C., Kong W., Cong L., Fu J., Zhang Z., Yu H., Wang Y., Jiang J. Abnormal metabolism of gut microbiota reveals the possible molecular mechanism of nephropathy induced by hyperuricemia. Acta Pharm. Sin. B. 2020;10:249–261. doi: 10.1016/j.apsb.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu J., Zhong X., Yan J., Zhou D., Qin D., Xiao X., Zheng Y., Liu Y. High-throughput sequencing analysis of intestinal flora changes in ESRD and CKD patients. BMC Nephrol. 2020;21 doi: 10.1186/s12882-019-1668-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guirong Y.E., Minjie Z., Lixin Y.U., Junsheng Y.E., Lin Y., Lisha S. Gut microbiota in renal transplant recipients, patients with chronic kidney disease and healthy subjects. Nan Fang Yi Ke Da Xue Xue Bao. 2018;38:1401–1408. doi: 10.12122/j.issn.1673-4254.2018.12.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamada N., Iwamoto C., Kano H., Yamaoka N., Fukuuchi T., Kaneko K., Asami Y. Evaluation of purine utilization by Lactobacillus gasseri strains with potential to decrease the absorption of food-derived purines in the human intestine. Nucleos Nucleot. Nucleic Acids. 2016;35:670–676. doi: 10.1080/15257770.2015.1125000. [DOI] [PubMed] [Google Scholar]
- 14.Yang H.-T., Xiu W.-J., Liu J.-K., Yang Y., Hou X.-G., Zheng Y.-Y., Wu T.-T., Wu C.-X., Xie X. Gut microbiota characterization in patients with asymptomatic hyperuricemia: probiotics increased. Bioengineered. 2021;12:7263–7275. doi: 10.1080/21655979.2021.1976897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu X., Ouyang S., Xie Y., Gong Z., Du J. Characterizing the gut microbiota in patients with chronic kidney disease. Postgrad. Med. 2020;132:495–505. doi: 10.1080/00325481.2020.1744335. [DOI] [PubMed] [Google Scholar]
- 16.Xi Y., Yan J., Li M., Ying S., Shi Z. Gut microbiota dysbiosis increases the risk of visceral gout in goslings through translocation of gut-derived lipopolysaccharide. Poultry Sci. 2019;98:5361–5373. doi: 10.3382/ps/pez357. [DOI] [PubMed] [Google Scholar]
- 17.Arman T., Lynch K.D., Goedken M., Clarke J.D. Sub-chronic microcystin-LR renal toxicity in rats fed a high fat/high cholesterol diet. Chemosphere. 2021;269 doi: 10.1016/j.chemosphere.2020.128773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lun H., Yang W., Zhao S., Jiang M., Xu M., Liu F., Wang Y. Altered gut microbiota and microbial biomarkers associated with chronic kidney disease. Microbiol. 2019;8 doi: 10.1002/mbo3.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wexler A.G., Goodman A.L. An insider's perspective: Bacteroides as a window into the microbiome. Nat Microbiol. 2017;2 doi: 10.1038/nmicrobiol.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao C., Fan B., Zhu J., Zhu N., Cao J.Y., Yang D.R. Association of gut microbiota and biochemical features in a Chinese population with renal uric acid stone. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.888883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y., Xu J., Wang X., Ren X., Liu Y. Changes of intestinal bacterial microbiota in coronary heart disease complicated with nonalcoholic fatty liver disease. BMC Genom. 2019;20:862. doi: 10.1186/s12864-019-6251-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J., Chen Y., Zhong H., Chen F., Regenstein J., Hu X., Cai L., Feng F. The gut microbiota as a target to control hyperuricemia pathogenesis: potential mechanisms and therapeutic strategies. Crit. Rev. Food Sci. Nutr. 2021:1–11. doi: 10.1080/10408398.2021.1874287. [DOI] [PubMed] [Google Scholar]
- 23.Hu X., Xie Y., Xiao Y., Zeng W., Gong Z., Du J. Longitudinal analysis of fecal microbiome and metabolome during renal fibrotic progression in a unilateral ureteral obstruction animal model. Eur. J. Pharmacol. 2020;886 doi: 10.1016/j.ejphar.2020.173555. [DOI] [PubMed] [Google Scholar]
- 24.Holecek M. Histidine in health and disease: metabolism, physiological importance, and use as a supplement. Nutrients. 2020;12 doi: 10.3390/nu12030848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durante W. Amino acids in circulatory function and health. Adv. Exp. Med. Biol. 2020;1265:39–56. doi: 10.1007/978-3-030-45328-2_3. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y., Yu W., Han D., Meng J., Wang H., Cao G. L-lysine ameliorates sepsis-induced acute lung injury in a lipopolysaccharide-induced mouse model. Biomed. Pharmacother. 2019;118 doi: 10.1016/j.biopha.2019.109307. [DOI] [PubMed] [Google Scholar]
- 27.Ko J.W., Kwon H.J., Seo C.S., Choi S.J., Shin N.R., Kim S.H., Kim Y.H., Kim J.C., Kim M.S., Shin I.S. 4-Hydroxycinnamic acid suppresses airway inflammation and mucus hypersecretion in allergic asthma induced by ovalbumin challenge. Phytother Res. 2020;34:624–633. doi: 10.1002/ptr.6553. [DOI] [PubMed] [Google Scholar]
- 28.Tan Y., Wang L., Gao J., Ma J., Yu H., Zhang Y., Wang T., Han L. Multiomics integrative analysis for discovering the potential mechanism of dioscin against hyperuricemia mice. J. Proteome Res. 2021;20:645–660. doi: 10.1021/acs.jproteome.0c00584. [DOI] [PubMed] [Google Scholar]
- 29.Li L., Li J., Cao H., Wang Q., Zhou Z., Zhao H., Kuang H. Determination of metabolic phenotype and potential biomarkers in the liver of heroin addicted mice with hepatotoxicity. Life Sci. 2021;287 doi: 10.1016/j.lfs.2021.120103. [DOI] [PubMed] [Google Scholar]
- 30.Jiao J., Zhang Y., Han P., Zhai S. A preliminary study on the value of intestinal flora in predicting major adverse cardiovascular and cerebrovascular events in patients with refractory hypertension. Comput. Math. Methods Med. 2022;2022 doi: 10.1155/2022/7723105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Zhang J., Feng D., Law H.K., Wu Y., Zhu G.H., Huang W.Y., Kang Y. Integrative analysis of gut microbiota and fecal metabolites in rats after prednisone treatment. Microbiol. Spectr. 2021;9 doi: 10.1128/Spectrum.00650-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gong H., Gao H., Ren Q., He J. The abundance of bifidobacterium in relation to visceral obesity and serum uric acid. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-17417-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this study has been deposited at SRA of NCBI under the accession number RJNA879265.