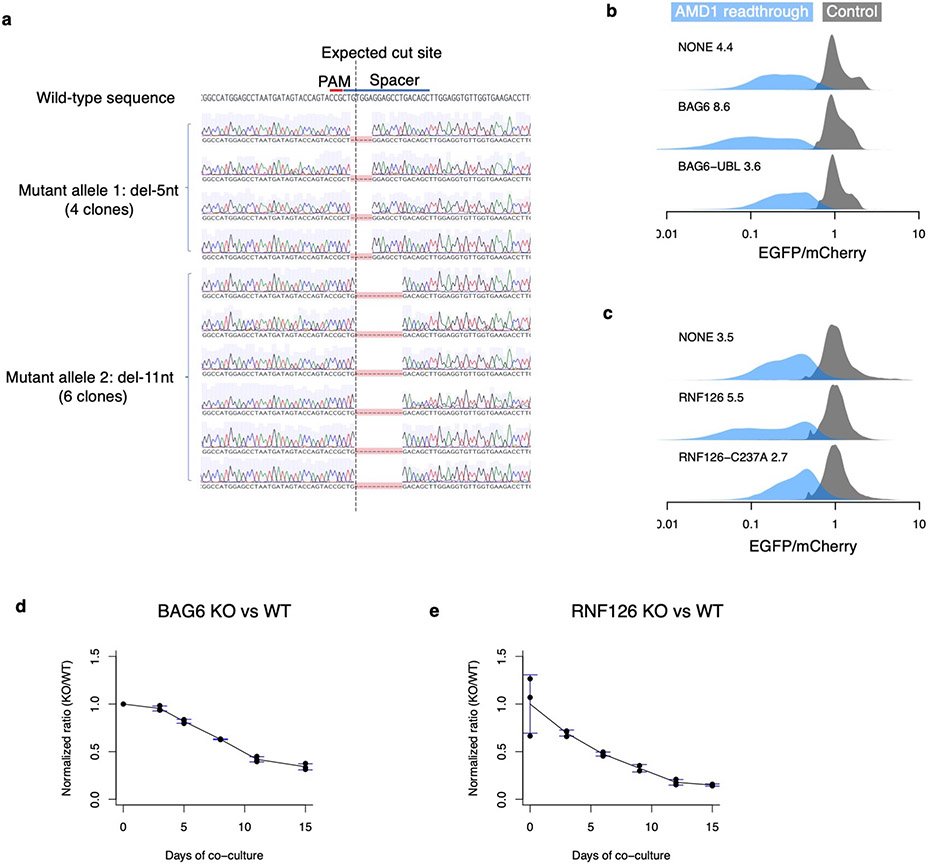
Extended Data Fig. 7 ∣. Characterization of the BAG6 KO cells and RNF126 KO cells.
a, Genotyping the BAG6 clonal knockout cell line. Sanger sequencing of 10 clones of PCR-amplified genomic DNA confirmed that the BAG6 KO cells contain a frameshift mutation in both alleles, one with a 5-nt deletion and the other with an 11-nt deletion around the expected Cas9 cut site. b, Re-expressing wild type BAG6 but not an inactive mutant missing the UBL domain for recruiting RNF126 (BAG6-UBL) partially reverses BAG6 KO phenotype as measured by the destabilization of AMD1 readthrough product. c, Same as b but comparing wild type RNF126 and an inactive mutant with a C237A mutation in the active site. d-e, Growth defect of BAG6 KO cells (d) and RNF126 KO cells (N=3 biologically independent samples) (e) revealed by competitive growth assays. KO cells and WT cells were mixed and co-cultured for 15 days and the relative cell numbers (KO/WT) at each time point was determined by decomposition of sanger sequencing traces as described in Methods. N=1 for day 0 of BAG6 and N=3 biologically independent samples for all other time points. Data are presented as mean values +/− s.d.