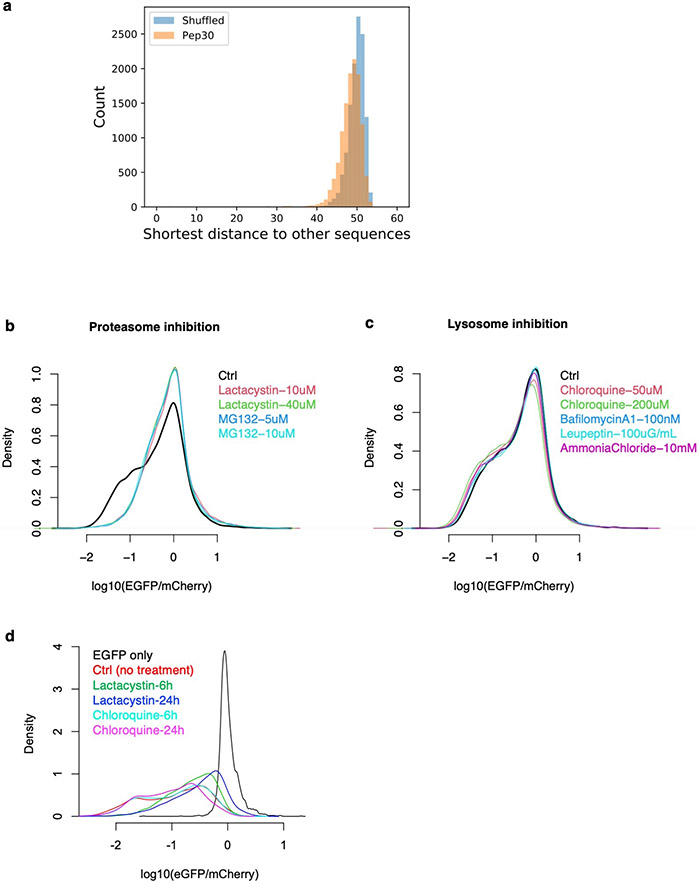
Extended Data Fig. 2 ∣. Characterization of the Pep30 library.
a, Sequence diversity in the Pep30 library. The pairwise hamming distance (number of nucleotides that are different) between any two sequences (of 90-nt) in the library was calculated. Subsequently for each sequence, we identify the shortest distance to any other sequence in the library. The result showed that the vast majority (98%) of Pep30 sequences are at least 40 nt (out of 90 nt) different from other sequences in the library, with a median distance of 48. This is very close the distribution when the Pep30 library sequences are shuffled (median: 50). The result indicated that our Pep30 library is nearly as diverse as one can get from entirely unrelated sequences. b-d, Effect of proteasome inhibition or lysosome inhibition on the Pep30 library. b, Pep30 cells were treated with proteasome inhibitors for 8 hours and then analyzed with flow cytometry. Ctrl: Pep30 cells without treatment. c, Same as (b) for multiple lysosome inhibitors. d, longer (24h vs. 6h) proteasome inhibition but not lysosome inhibition resulted in more rescue.