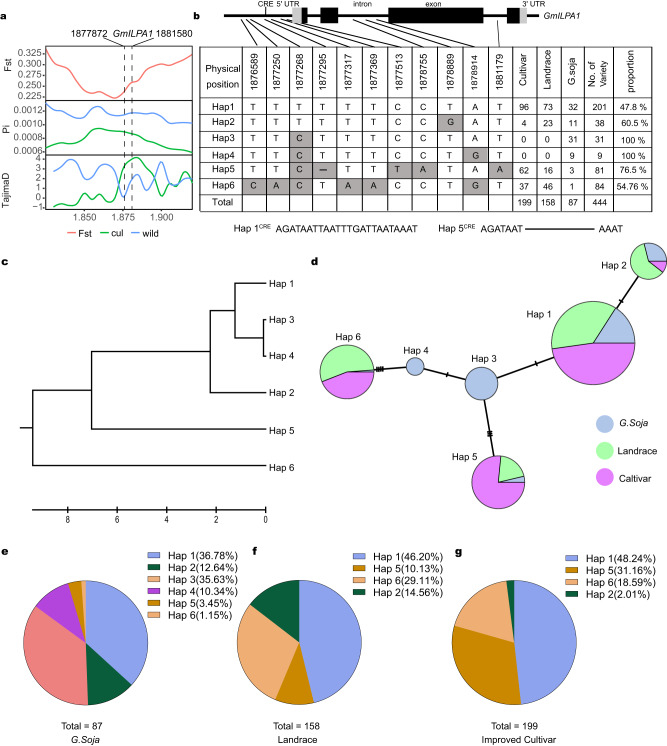
Fig. 6. Haplotype analysis and origin of GmILPA1 in soybean accessions from different populations of China.
a FST, nucleotide diversity, and Tajima’s D values over the genomic region containing the GmILPA1 locus (~100 kb) between wild and cultivar soybean germplasms. b GmILPA1 haplotypes in natural populations. Top, schematic diagram of the GmILPA1 gene structure; gray, untranslated regions (UTRs); black, exons; black lines, promoter and introns. The CRE in the promoter region represents a light-responsive cis-regulatory element; –, 13-bp deletion. Bottom, GmILPA1 polymorphism across accessions relative to the Williams 82 reference genome (Hap1). The number of varieties for each haplotype (Hap1–6) is shown to the right. The cultivated soybean Hedou 12 belongs to Hap5. c Evolutionary relationship of the GmILPA1 six haplotypes. d Haplotype origins of GmILPA1 as shown by the median-joining method. Circle size is proportional to the number of accessions, while circle colors represent the different soybean groups: blue, wild soybean (S. soja); green, landraces; magenta, cultivars. e–g Pie charts representing the distribution of each haplotype in wild soybeans, landraces, and improved cultivars.