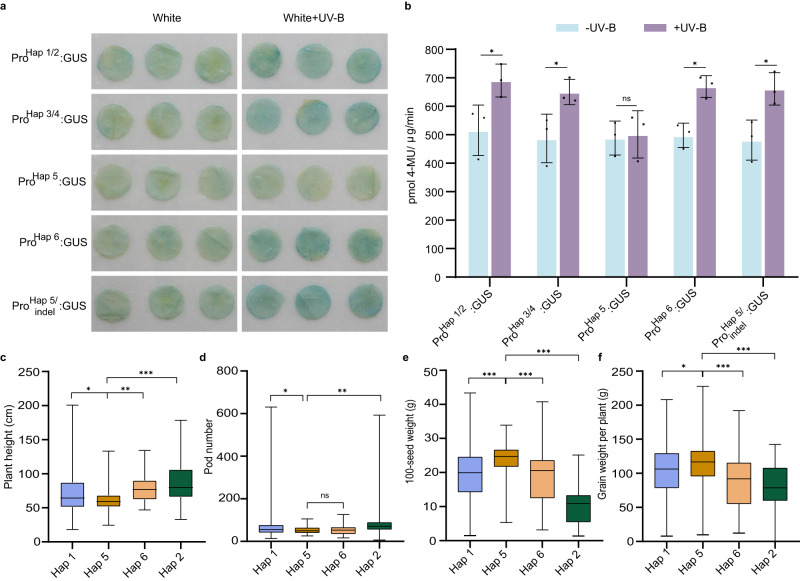
Fig. 7. Different Haplotypes of GmILPA1 confers variation of plant height in response to UV-B in cultivars.
a GUS activity detected by histochemical staining in Nicotiana benthamiana leaves transiently expressing the GUS reporter gene under the control of GmILPA1Hap1- GmILPA1Hap6 and the GmILPA1Hap5 promoter with indel. For UV-B treatment, N. benthamiana were treated with UV-B (1.5 μmol m−2 s−1) for 12 h before histochemical staining. b GUS activity from N. benthamiana leaves transiently expressing GUS under the control of GmILPA1Hap1- GmILPA1Hap6 and the GmILPA1Hap5 promoter, as measured by fluorometric assay and expressed as pmol 4-methylumbelliferone μg–1 protein min–1, (n = 3 independent experiments, P = 0.0457 in ProHap1/2: GUS, P = 0.0425 in ProHap3/4: GUS, P = 0.0413 in ProHap6: GUS and P = 0.0263 in ProHap5/indel: GUS). Data are presented as mean values ± SD, Student’s t-test was used for the significance test, ns, not significant, *P < 0.05. c–f Summary of major agronomic traits plants with the GmILPA1Hap1, GmILPA1Hap2, GmILPA1Hap5or GmILPA1Hap6 allele. (P = 0.0409 in Hap 1 and Hap 5, P = 0.0088 in Hap 6 and Hap 5, P = 0.0006 in Hap 2 and Hap 5) (c), (P = 0.0233 in Hap 1 and Hap 5, P = 0.0017 in Hap 2 and Hap 5) (d), (P = 2.61 × 10−6 in Hap 1 and Hap 5, P = 3.57 × 10−6 in Hap 6 and Hap 5, P = 8.95 × 10−21 in Hap 2 and Hap 5) (e), (P = 0.0143 in Hap 1 and Hap 5, P = 5.35 × 10−6 in Hap 6 and Hap 5, P = 1.33 × 10−6 in Hap 2 and Hap 5) (f). n = 179 for GmILPA1Hap1, n = 33 for GmILPA1Hap2, n = 76 for GmILPA1Hap5, n = 81 for GmILPA1Hap6. The box indicates the range from lower to upper quartiles, and the bar ranges the minimum to maximum observations; the significance of the difference is calculated by comparison with Hap 5 with a one-way ANOVA analysis–Tukey comparison and the columns labeled without the same alphabet are significantly different (P < 0.05, two-sided). *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant. Source data are provided as a Source Data file.