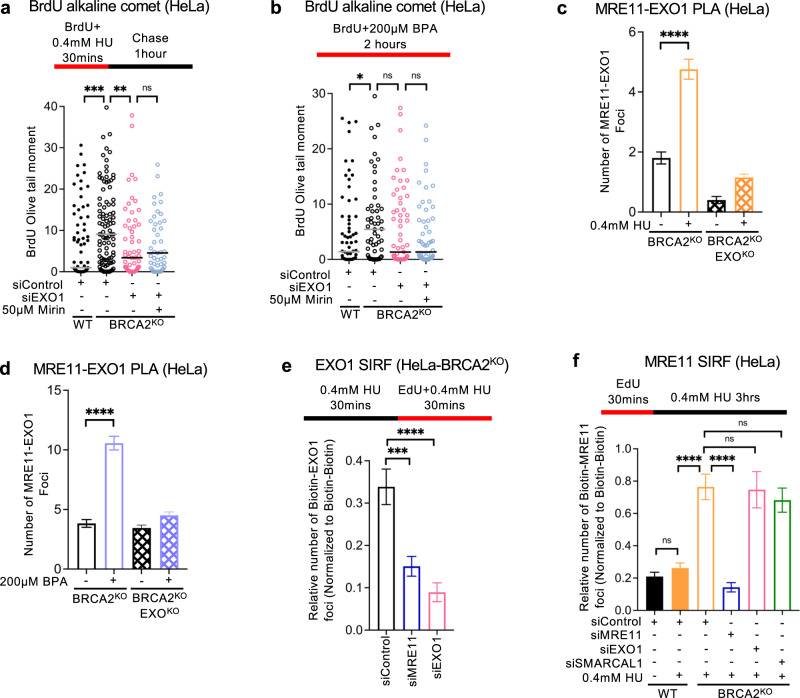
Fig. 5. Regulation of EXO1 and MRE11 recruitment to nascent strand ssDNA gaps.
a, b BrdU alkaline comet assays showing that treatment of EXO1-depleted HeLa-BRCA2KO cells with mirin does not further suppress the accumulation of ssDNA gaps induced by HU (a) or BPA (b) exposure. At least 40 nuclei were quantified for each condition. The median values are marked on the graph. Asterisks indicate statistical significance (Mann–Whitney, two-tailed). c, d PLA assays showing that EXO1 and MRE11 co-localize upon treatment with 0.4 mM HU (c) or 200 µM BPA (d) for 3 h. EXO1 deletion is used as control to confirm the specificity of the PLA signals observed. At least 75 cells were quantified for each condition. Bars indicate the mean values, error bars represent standard errors of the mean, and asterisks indicate statistical significance (t test, two-tailed, unpaired). e, f EXO1 (e) and MRE11 (f) SIRF experiments showing the differential impact of the loss of each of these nucleases on the recruitment to nascent DNA of the other nuclease, upon exposure of HeLa-BRCA2KO cells to 0.4 mM HU. Loss of MRE11 suppressed EXO1 binding to nascent DNA, while EXO1 depletion did not affect MRE11 recruitment under these conditions. Depletion of MRE11 or EXO1 respectively is used as control to confirm the specificity of the SIRF signals observed. At least 45 cells were quantified for each condition. Bars indicate the mean values, error bars represent standard errors of the mean, and asterisks indicate statistical significance (t test, two-tailed, unpaired). Schematic representations of the assay conditions are shown at the top. Source data are provided as a Source data file.