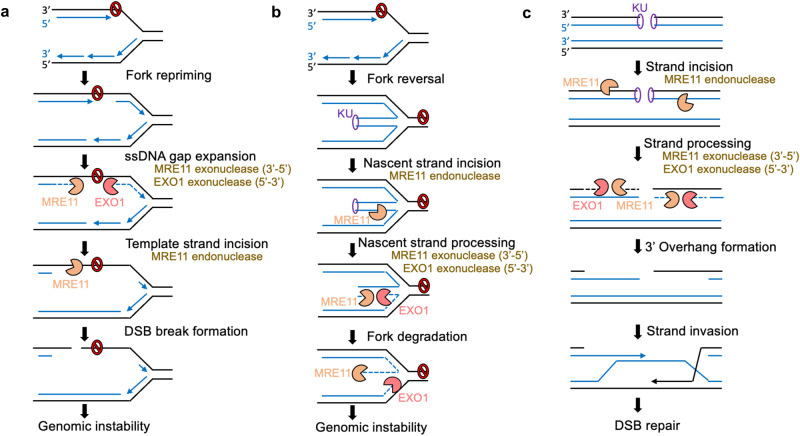
Fig. 7. Schematic representation of the proposed models.
The sequential activities of EXO1 and MRE11 exonuclease activities and or MRE11 endonuclease activity in ssDNA gap formation (a), degradation of reversed forks (b), and end resection during DSB repair by HR (c) are shown. In all cases, the MRE11 3’−5’ exonuclease activity and the EXO1 5’−3’ exonuclease activity bidirectionally expand a ssDNA nick or short gap. In ssDNA gap processing (a), the MRE11 endonuclease cleaves the template strand to cause a DSB subsequent to gap expansion. In fork degradation (b) and DSB end resection (c), the DSB ends are protected by binding of the KU complex, which prevents the direct engagement of exonuclease activities. The endonuclease activity of MRE11, which is recruited to dsDNA in the proximity of the KU-bound DSB end, is required to create the initial nick extended by the exonuclease activities or EXO1 and MRE11. MRE11-mediated exonucleolytic degradation towards the DSB end results in removal of the KU complex, while EXO1 catalyzes long-range resection in the opposite direction (moving away from the DSB end).