Figure 5. Fluorescence HPLC of released N-glycans defines a Fuc3 species enriched on SVs.
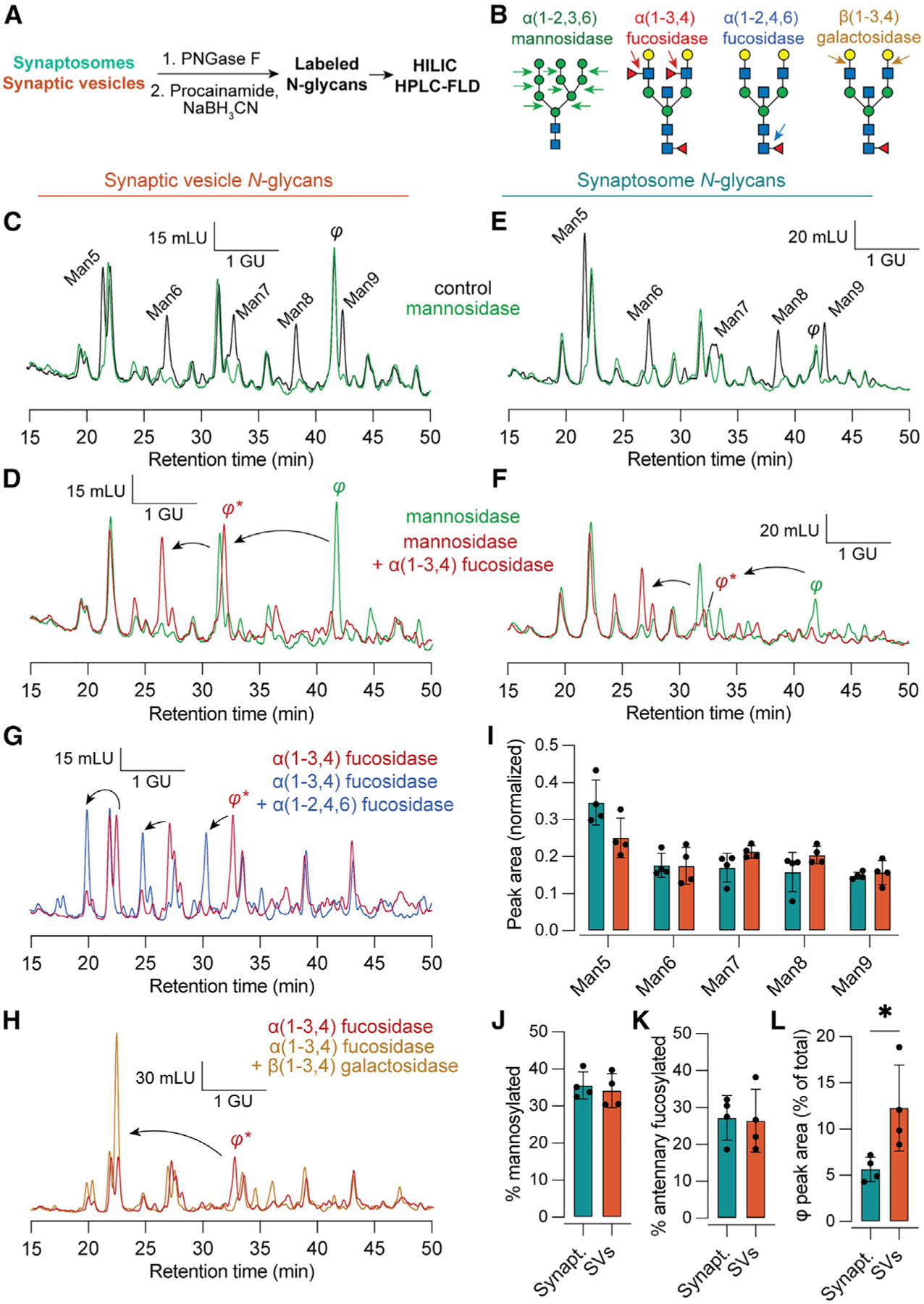
(A) Experimental scheme.
(B) Illustration of cleavage sites for exoglycosidases used for this figure. α(1–2,3,6) mannosidase cleaves mannose residues from mannosylated N-glycans. α(1–3,4) fucosidase removes only antennary fucose but not the core fucose, which is sensitive to α(1–2,4,6) fucosidase. β(1–3,4) galactosidase removes antennary galactose residues.
(C) Representative chromatograms of procainamide-labeled SV N-glycans under control conditions and treated with mannosidase. The identity of the mannose series was established using an authentic Man5 standard. Mannosidase-sensitive peaks appear in black. A mannosidase-insensitive peak eluting between Man8 and Man9 is labeled φ. GU, glucose units, determined via procainamide-labeled glucose homopolymer standard; mLU, milli-luminance units.
(D) The addition of α(1–3,4) fucosidase causes φ to migrate ~1.9 GU, indicating the presence of two antennary fucoses.
(E and F) As in (C) and (D) but for synaptosome N-glycans. While a peak corresponding to φ is observed in both sample types, this species is relatively more abundant in SVs.
(G) After treatment with α(1–3,4) fucosidase, φ remains sensitive to α(1–2,4,6) fucosidase, consistent with α(1–6) core fucosylation and a total of 3 fucoses.
(H) φ was also sensitive to β(1–3,4) galactosidase, which caused an additional ~2 GU shift.
(I–K) The distribution of peak areas among mannosylated glycans, as well as the total contribution from mannosylated and antennary fucosylated glycans, was similar in synaptosomes and SVs.
(L) The peak corresponding to φ made a significantly larger contribution to SV versus synaptosome glycans (p = 0.02, Mann-Whitney test), demonstrating enrichment of this Fuc3 glycan in SVs. Data are represented as mean ± SD from four biological replicates.
See also Figure S5, which contains additional raw HPLC data used to characterize peaks corresponding to Man5–9 and φ.
