Figure 1. Engineering of ultrasensitive and highly responsive lactate sensors.
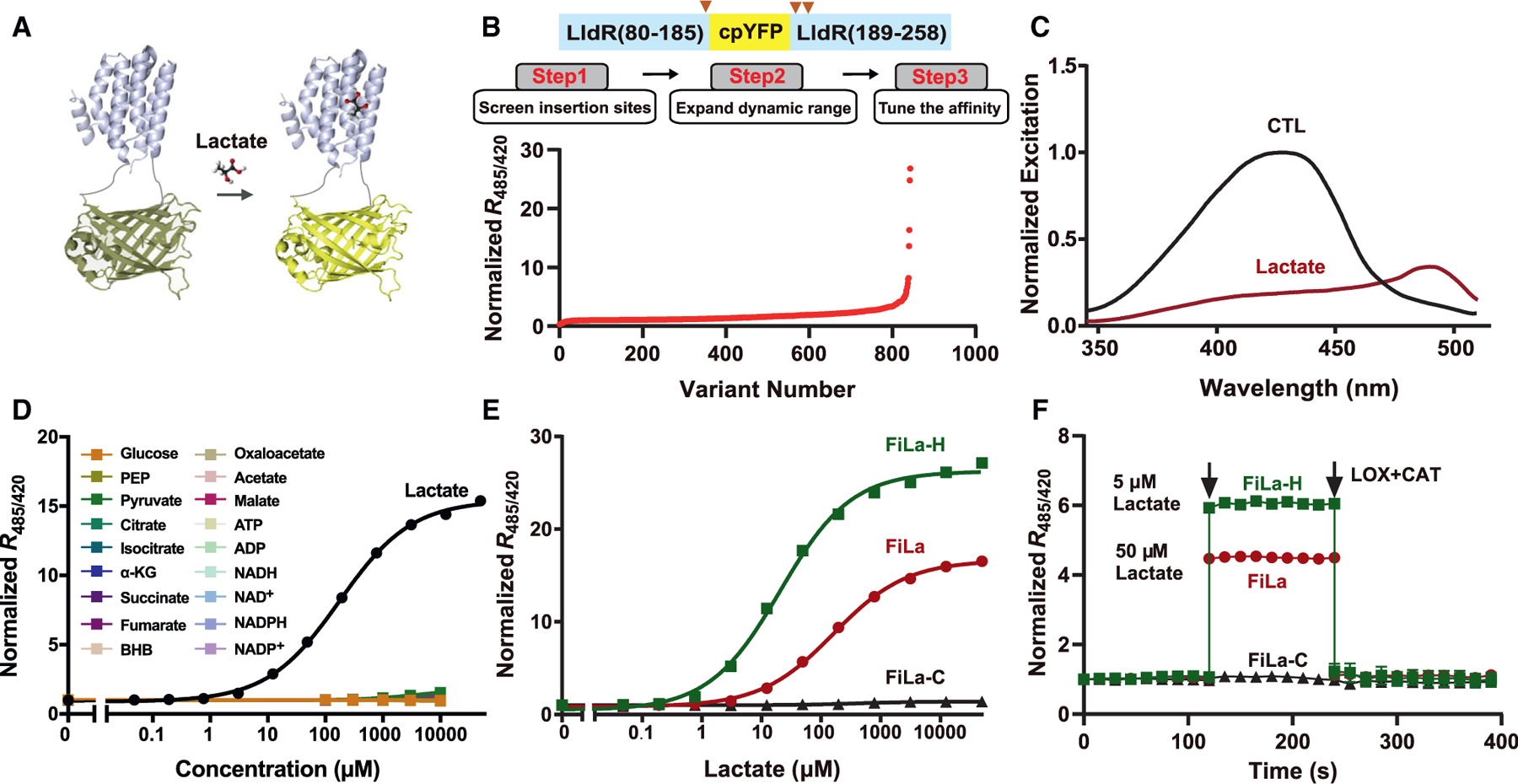
(A) Schematic representation of the lactate sensor FiLa. The fluorescent protein cpYFP was inserted into a monomer of the lactate-binding bacterial protein LldR. Binding of lactate induces changes in protein conformation and fluorescence.
(B) Engineering of ultrasensitive and highly responsive lactate sensors. A total of 843 variants was screened, including cpYFP insertion sites, truncated variants, random mutants, and site-saturation mutants.
(C) Excitation spectra of purified FiLa in the control condition (black) and saturated with lactate (dark red). The excitation spectrum recorded at an emission wavelength of 530 nm has maxima around 425 and 490 nm. Data are normalized to the peak intensity in the control condition.
(D) Fluorescence response of FiLa at the indicated concentration of lactate or other metabolites. Data are normalized to the initial value (n = 3). PEP, phosphoenolpyruvate; α-KG, α-ketoglutarate; BHB, β-hydroxybutyrate.
(E) Lactate titration curves of FiLa, FiLa-H, and FiLa-C sensors. Data are normalized to the initial value (n = 3).
(F) Kinetics of fluorescence response of purified FiLa and FiLa-H protein to 50 μM lactate and 5 μM lactate, respectively. Lactate was subsequently decreased by the addition of 100 μM lactate oxidase (LOX) and 500 U/mL catalase (CAT). Data are normalized to the initial value (n = 3). Data are the mean ± SEM (D–F). See also Figure S1 and Tables S1 and S2.
