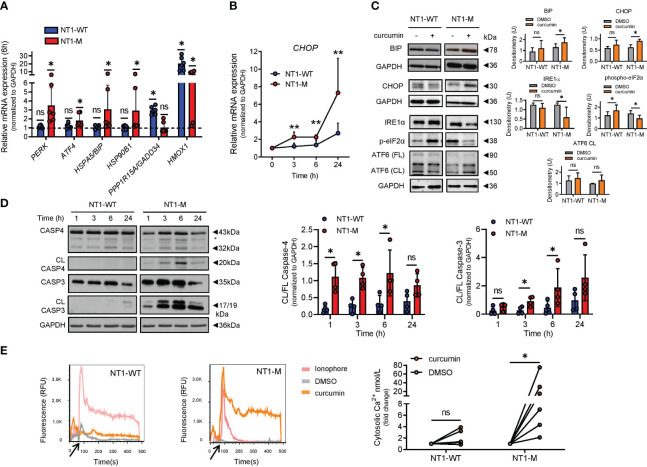
Figure 2.
UPR/ER stress response differs between NT1-M and NT1-WT CLL cells after curcumin treatment. Primary CLL cells (2 × 106 cells/ml) were incubated with 15 µM curcumin or vehicle DMSO at 0.05%. (A) Bar graphs showing the transcriptional expression of ER UPR markers PERK, ATF4, BIP/HSPA5, and HSP90B1 and the stress-overcoming genes GADD34 and HMOX1, in NT1-WT and NT1-M cells assessed via qPCR (N = 6, per group), after 6 h of incubation. mRNA levels were normalized to GAPDH and are represented as fold change using cells incubated with DMSO as a reference set to 1 (dashed line). (B) Kinetics of mRNA expression of the pro-apoptotic transcription factor CHOP after curcumin treatment (N = 8, per group). (A, B) Data are presented as mean ± SD. *P< 0.05; ns, not significant as determined by Wilcoxon paired test (A). **P< 0.01 as determined by Mann–Whitney unpaired test. (C) Representative Western blots (left) of BiP, CHOP, IRE1α, phosphorylated eIF2α (p-eIF2α), and full-length (FL) and cleaved (CL) ATF6 after 24-h treatment with curcumin or DMSO and bar graphs (right) with densitometry data of NT1-WT and NT1-M cells (N = 6, per group). (D) Representative Western blots (left) of caspase 4 and caspase 3 cleavage at the indicated time points and bar graphs (right) with data points of densitometric analysis of CL/FL caspase 4 and CL/FL caspase 3 in NT1-WT and NT1-M cells (N = 4, per group). Data are presented as mean ± SD. *P< 0.05; ns, not significant as determined by Mann–Whitney unpaired test. (E) Cytosolic Ca2+ measurement of CLL cells loaded with Fluo-4. Left, relative fluorescence units (RFU) from Fluo-4 were recorded during a 60-s period in basal conditions (black arrow) and after the addition of calcium ionophores (2 μM), DMSO, or curcumin, for 420 s. Right, dot-and-line diagram of the fold change of cytosolic Ca2+ levels (nmol/L) using cells incubated with DMSO as a reference set to 1, in NT1-WT or NT1-M (N = 6, per group). *P< 0.05; ns, not significant as determined by Wilcoxon paired test.