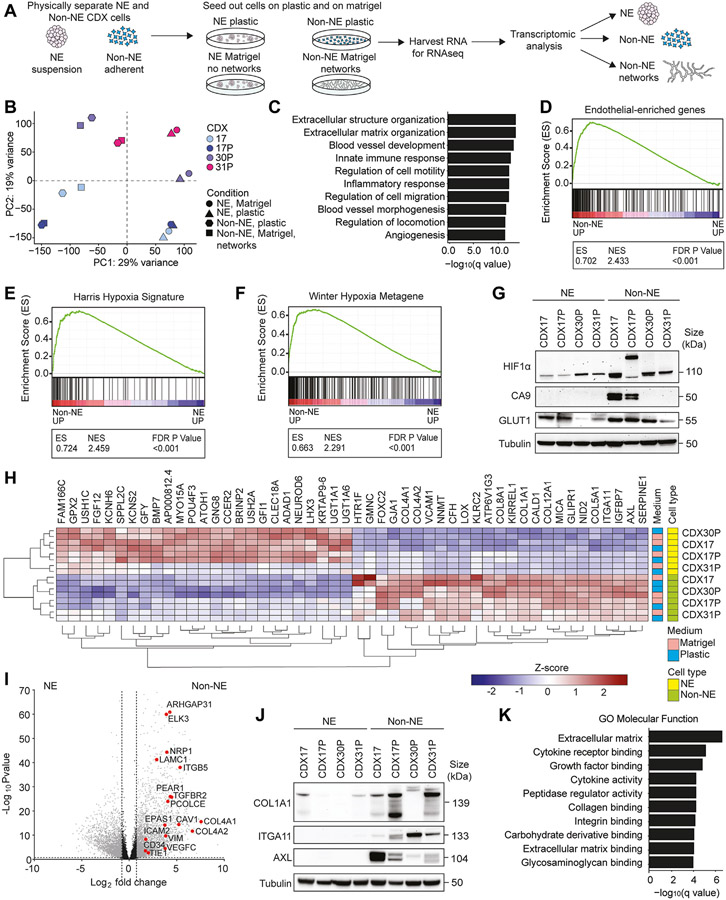
Figure 4.
Transcriptomic analysis of network-forming CDX cells. (A) Workflow for generation of CDX suspension NE and adherent non-NE cells that were physically separated and cultured on plastic and Matrigel. From these samples, RNA was isolated for RNAseq followed by transcriptomic analysis. (B) Principal component analysis of CDX (CDX17, light blue, CDX17P, dark blue, CDX30P, purple, CDX31P, pink) NE and non-NE cell transcriptomes cultured on plastic and Matrigel. Each symbol represents an individual replicate: circles, NE cells on Matrigel, triangles, NE cells on plastic, hexagons, non-NE cells on plastic, squares, non-NE cells on Matrigel (network-forming) per CDX model. (C) GSEA of biological processes up-regulated in CDX non-NE cells compared with CDX NE cells. (D) GSEA of endothelial enriched genes47 in CDX NE and non-NE cell transcriptomes. The endothelial gene set was refined to remove any genes that are expressed in mesenchymal cells.48 (E) GSEA of Harris hypoxia signature52 in CDX NE and non-NE cell transcriptomes. (F) GSEA of Winter hypoxia metagene signature53 in CDX NE and non-NE cell transcriptomes. (G) Representative immunoblots in CDX NE and non-NE cell lysates. There are 2 to 3 independent replicate tumors per CDX. Tubulin loading control was run subsequently for each marker illustrated on the same blot. (H) Heatmap of the top 25 up-regulated and down-regulated genes in CDX non-NE cells (green) compared with NE cells (yellow), cultured on either plastic (blue) or Matrigel (pink). (I) Volcano plot of differentially expressed genes in CDX non-NE cells compared with NE cells. Significant (fold change >1, −log(qvalue) >1) genes in gray. (J) Representative immunoblots of CDX NE and non-NE cell lysates. There are 2 to 3 independent replicate tumors per CDX. Tubulin loading control was run subsequently for each marker illustrated on the same blot (K) GO molecular functions up-regulated in CDX non-NE cells compared with CDX NE cells. For GSEA in C, D, E, F, and K, NE and non-NE cells grown on plastic and Matrigel were combined and treated as technical replicates because there were no significant transcriptomic changes identified between these culture conditions. CDX, circulating tumor cell-derived explant; GO, gene ontology; GSEA, gene set enrichment analysis; NE, neuroendocrine; RNAseq, RNA sequencing.