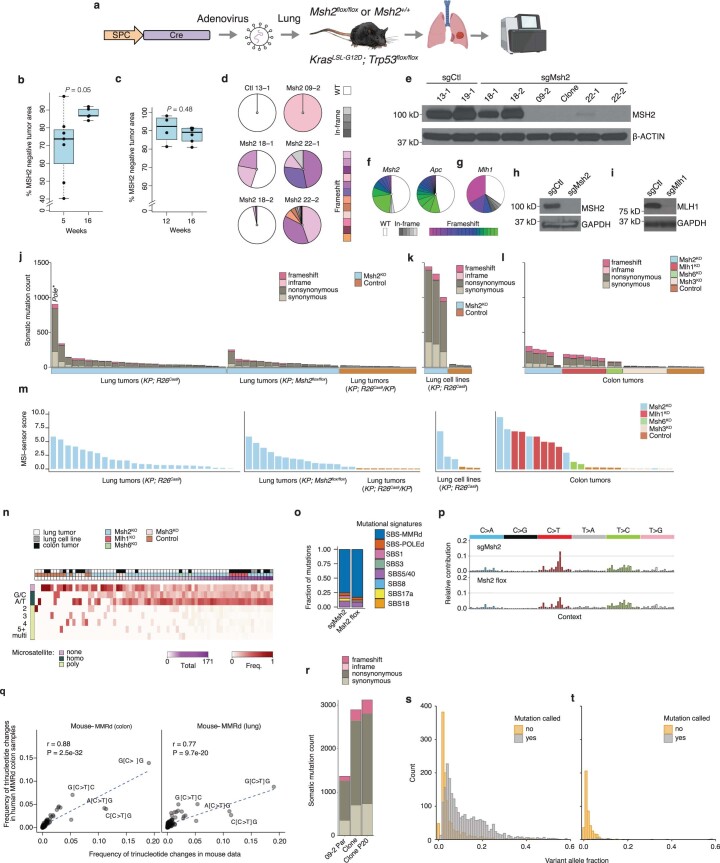
Extended Data Fig. 1. Validation of in vivo DNA mismatch repair gene knockout.
(a) Schematic of KrasLSL-G12D; Trp53flox/flox; Msh2floxl/flox (KPM) lung tumor model. (b-c) Percent lung tumor area negative for MSH2 by IHC at 5- and 16-weeks post-initiation of KrasLSL-G12D; Trp53flox/flox; R26LSL-Cas9 (KPC) animals with sgMsh2 lentivirus (N = 7 5-week and 5 16-week animals) (b) and 12- and 16-weeks post-initiation of KPM animals (N = 4 12-week and 6 16-week animals) (c). (d) Deep sequencing of sgMsh2-targeted locus in cell lines derived from 16–20-week sgCtl- and sgMsh2-targeted lung tumors. WT = wild-type. (e) Western blot of MSH2 expression in lung tumor cell lines, experimentally replicated three times. Clone = 09-2 single cell clone. (f-g) Deep sequencing of Msh2, Apc (f) and Mlh1 (g) loci targeted in autochthonous colon tumors, representative of 5 sgMsh2- and 6 sgMlh1-targeted tumors. (h-i) Western blots of MSH2 (h) and MLH1 (i) expression in one organoid line each derived from sgCtl-, sgMsh2-, and sgMlh1-targeted colon tumors, experimentally replicated twice. (j-l) Total somatic single nucleotide variants (SNVs) and insertions/deletions (indels) within exome of autochthonous lung tumors (j), cell lines (k), and autochthonous colon tumors (l). Pole* = Pole S415R mutation. (m) MSIsensor-pro scores for tumors in (j-l). (n) Frequency of indels across DNA microsatellite contexts, including exonic and intronic mutations. Samples were ordered by total indels. Homo = homopolymer repeats ≥ 4 bases, 2-5 + = microsatellites with motifs of 2-5+ bases, and multi = microsatellites with multiple repetitive motifs. (o-p) COSMIC mutational signature decomposition of 15 KPM and 26 KPC (sgMsh2) lung tumors (o) based on frequencies of the 96 possible SNVs classified by trinucleotide context (p). SBS_MRD = DNA mismatch repair deficiency (MMRd) signature. (q) Pearson correlation of the fraction of mutations detected in each trinucleotide context between human and mouse MMRd datasets. (r) Total somatic SNVs/indels in exome of 09-2 lung tumor cell line and early- and 20-passage 09-2 single cell clone. (s-t) Total mutations in early-passage clone that are also supported in sequencing reads of 09-2 parental line (s) and an unrelated MMRp control line, 13-1 (t), with variant allele fraction on x-axis. Grey = official mutation calls; gold = not called. Significance in (b-c) was assessed by Wilcoxon Rank Sum test.