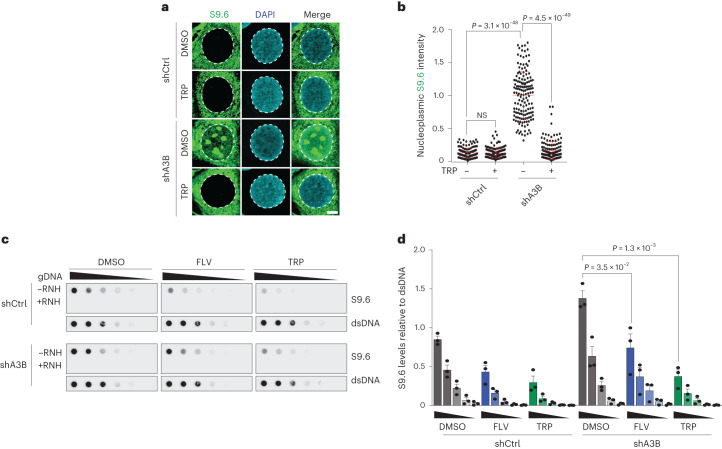
Fig. 4. A3B-regulated R-loops are transcription-dependent.
a,b, IF images (a) and quantification (b) of U2OS shCtrl and shA3B cells treated with TRP (1 μM, 4 h) and subsequently stained with S9.6 (green) and DAPI (blue; representative images; 5 μm scale; n = 3 independent experiments with >100 nuclei per condition; red bars represent mean ± s.e.m.; P value by Mann–Whitney test). c,d, S9.6 dot-blot analysis of a genomic DNA dilution series ± RNH from U2OS shCtrl or shA3B cells treated with TRP (1 μM, 4 h) or FLV (1 μM, 1 h; representative images); parallel dsDNA dot blots provided a loading control (c). Quantification was normalized to the most concentrated shCtrl/DMSO signal (n = 3 independent experiments; mean ± s.e.m.; P value by two-tailed unpaired t-test) (d). TRP, triptolide; FLV, flavopiridol.