Correction to: Scientific Reports 10.1038/s41598-023-42037-w, published online 16 September 2023
The original HTML version of this Article contained errors in Figures 1 to 5, where the dendrograms were not displayed correctly.
The original Figures 1, 2, 3, 4 and 5 and their accompanying legends appear below.
Figure 1.
Genome-wide DNA methylation analysis of CTC-MCC-41 with respect to HT29 primary tumor cells. (A) Schematic flowchart used to identify significant differentially methylated CpGs in CTC-MCC-41 compared to HT29. (B) Principal component analysis (PCA) of DNA methylation data obtained in CTC-MCC-41 and HT29 cells. (C) Scatter plot representing the mean normalized levels of DNA methylation (β-values) in CTC-MCC-41 and HT29 cells. Dots in red show significantly differentially methylated CpGs. (D) Hierarchical clustering heatmap of the 10,000 most differentially methylated CpGs (FDR adjusted p value < 0.05) between CTC-MCC-41 and HT29. Heatmap shows three different passages (P) of CTC-MCC-41 (P12, P13 and P14) and HT29 (P2, P3 and P4). (E, F) Description of the 188,185 differentially methylated CpGs (DMCpGs) found in CTC-MCC-41 with respect to HT29 according to (E) chromosome location and methylation status and (F) CpG context, gene location and transcription factor-binding sites (TFBS). QC quality control, FDR false discovery rate, CpGI CpG island, HypoM hypomethylated, HyperM hypermethylated.
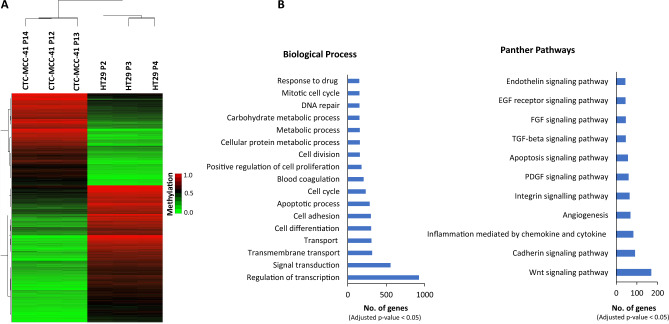
Figure 2.
DNA methylation profiles of CpGIs and shore regions of gene promoters in CTC-MCC-41 compared to HT29 primary tumor cells. (A) Hierarchical clustering heatmap of the 10,000 most differentially methylated CpGs (FDR adjusted p value < 0.05) in CTC-MCC-41 with respect to HT29 and located at CpGIs and shore regions of gene promoters. Heatmap shows three different passages (P) of CTC-MCC-41 (P12, P13 and P14) and HT29 (P2, P3 and P4). (B) Gene Ontology (GO) analysis representing some of the most cancer-relevant biological processes and Panther pathways based on the 10,000 most differentially methylated CpGs of CTC-MCC-41 compared to HT29 and located at CpGIs and shore regions of gene promoters.
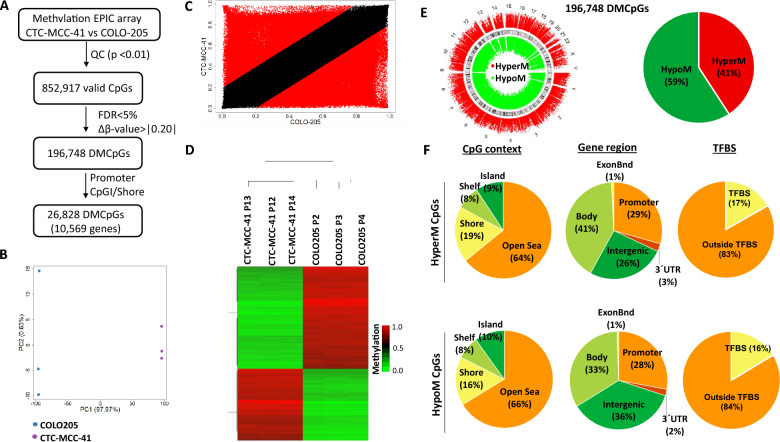
Figure 3.
Genome-wide DNA methylation analysis of CTC-MCC-41 with respect to COLO205 metastatic tumor cells. (A) Schematic flowchart used to identify significantly differentially methylated CpGs in CTC-MCC-41 compared to COLO205. (B) Principal component analysis (PCA) of DNA methylation data obtained for CTC-MCC-41 and COLO205 cells. (C) Scatter plot representing the mean normalized levels of DNA methylation (β-values) in CTC-MCC-41 and COLO205 cells. Dots in red show significantly differentially methylated CpGs. (D) Hierarchical clustering heatmap of the 10,000 most differentially methylated CpGs (FDR adjusted p value < 0.05) between CTC-MCC-41 and COLO205. Heatmap shows three different passages (P) of CTC-MCC-41 (P12, P13 and P14) and COLO205 (P2, P3 and P4). (E, F) Description of the 188,185 differentially methylated CpGs (DMCpGs) found in CTC-MCC-41 with respect to COLO205 according to (E) chromosome location and methylation status and (F) CpG context, gene location and transcription factor-binding sites (TFBS). QC quality control, FDR false discovery rate, CpGI CpG island, HypoM hypomethylated, HyperM hypermethylated.
Figure 4.
DNA methylation patterns of CpGIs and shore regions of gene promoters in CTC-MCC-41 compared to COLO205 metastatic tumor cells. (A) Hierarchical clustering heatmap of the 10,000 most differentially methylated CpGs (FDR adjusted p value < 0.05) in CTC-MCC-41 with respect to COLO205 and located at CpGIs and shore regions of gene promoters. Heatmap shows three different passages (P) of CTC-MCC-41 (P12, P13 and P14) and COLO205 (P2, P3 and P4). (B) Gene Ontology (GO) analysis representing some of the most cancer-relevant biological processes and Panther pathways based on the 10,000 most differentially methylated CpGs of CTC-MCC-41 compared to COLO205 and located at CpGIs and shore regions of gene promoters.
Figure 5.
DNA methylation signature of CTC-MCC-41 with respect to colorectal primary and metastatic tumor cells. (A) Hierarchical clustering heatmap with the 17,827 differentially methylated CpGs (FDR adjusted p value < 0.05) in CTC-MCC-41 cells compared to HT29 and COLO205 cells, representing primary and metastatic tumor cells, respectively. (B) Hierarchical clustering heatmap of the 9,949 differentially methylated CpGs (FDR adjusted p value < 0.05) in CTC-MCC-41 compared to several colorectal primary (HT29, Caco2, HCT116, RKO) and metastatic tumor cells (COLO205 and SW620). Heatmaps show three different passages (P) of CTC-MCC-41 (P12, P13 and P14), HT29 (P2, P3 and P4), and COLO205 (P2, P3 and P4).
These errors have now been corrected in the HTML version of the Article; the PDF version was correct from the time of publication.
Contributor Information
Rafael López-López, Email: rafael.lopez.lopez@sergas.es.
Catherine Alix-Panabières, Email: c-panabieres@chu-montpellier.fr.
Angel Díaz-Lagares, Email: angel.diaz.lagares@sergas.es.