Abstract
Aggregation and fibrillization of transthyretin (TTR) is a fatal pathogenic process that can cause cardiomyopathic and polyneuropathic diseases in humans. Although several therapeutic strategies have been designed to prevent and treat related pathological events, there is still an urgent need to develop better strategies to improve potency and wider applicability. Here, we present our study demonstrating that 3-iodothyronamine (T1AM) and selected thyronamine-like compounds can effectively prevent TTR aggregation. T1AM is one of the thyroid hormone (TH) metabolites, and T1AM and its analogs, such as SG2, SG6, and SG12, are notable molecules for their beneficial activities against metabolic disorders and neurodegeneration. Using nuclear magnetic resonance (NMR) spectroscopy and biochemical analysis, we confirmed that T1AM analogs could bind to and suppress acid-induced aggregation of TTR. In addition, we employed computational approaches to further understand the detailed mechanisms of the interaction between T1AM analogs and TTR. This study demonstrates that T1AM analogs, whose beneficial effects against several pathological processes have already been proven, may have additional benefits against TTR aggregation and fibrillization. Moreover, we believe that our work provides invaluable insights to enhance the pleiotropic activity of T1AM and structurally related analogs, relevant for their therapeutic potential, with particular reference to the ability to prevent TTR aggregation.
Keywords: Thyronamine, Thyroid hormone analog, Transthyretin amyloidosis, Molecular dynamics simulation, Nuclear magnetic resonance spectroscopy
Graphical Abstract
Highlights
-
•
Analogs of thyroid hormone derivatives are promising multifunctional drug molecules.
-
•
T1AM analogs are the effective binders of transthyretin.
-
•
T1AM analogs suppress aggregation and amyloidosis of transthyretin.
-
•
T1AM analogs are promising therapeutic candidates for transthyretin-related diseases.
1. Introduction
Thyroid hormones (THs) are essential signaling molecules that modulate diverse physiological and pathological processes [1], [2]. Upon secretion from the thyroid, the predominant forms of THs are L-thyroxine (T4) and 3,3′,5-triiodo-L-thyronine (T3), of which T3 is the active form that mediates most of the TH-related physiological processes. In 2004, however, it was discovered that in human blood and tissues, there is another active endogenous molecule whose physiological activity is mostly antagonistic to T3, and subsequent mass-spectrometric analyses determined its chemical structure as a mono-iodine-containing thyronamine, 3-iodothyronamine (T1AM) [3]. In contrast to the classical signaling targets of T3, such as the nuclear thyroid hormone receptor, the major target of T1AM was identified as trace amine-associated receptor families (such as TAAR1), thus mediating different signaling pathways from those of T3 [4].
The discovery of T1AM and its distinctive signaling pathways has attracted considerable attention owing to its potential as a novel therapeutic intervention for various human diseases [1], [5], [6]. In particular, several research groups have identified that T1AM and its analogs are effective not only for metabolic disorders, but also for neurodegenerative diseases (NDDs) [5], [7], [8]. A series of reports have demonstrated that the neuroprotective effects of T1AM and its analogs are mediated by an increase in ERK1/2 phosphorylation and transcription factor c-fos expression [5], by modulation of SIRT6 and autophagy induction [7], or by interplay with the histaminergic system [8], [9]. Notably, Chiellini et al. developed a 3-methylbiaryl-methane analog of T1AM, SG2, and its potential oxidative metabolite SG6, and found that they exhibited therapeutic activities against NDDs [5], [10], [11]. In addition, an aspect of considerable importance in the development of drugs intended for the treatment of CNS disorders is to know the effective delivery of these agents across the blood brain barrier (BBB). In a recent study, di Leo et al. [6] investigated the permeability of an in vitro model of BBB to T1AM, showing compelling evidence that T1AM was able to efficiently cross the BBB.
One of the advantageous features of TH analogs as drugs is their multifunctionality, which simultaneously target various physiological processes. T1AM can target multiple receptors and channels, for example, TAAR families (TAAR1, TAAR5, and TAAR8), ADRA2A, TRPM8, TRPV1, and F0F1-ATPase, implying that similar multi-functionality is also expected for T1AM analogs [11]. However, there are several TH transporter proteins, such as thyroxine-binding globulin, transthyretin (TTR), and albumin [12], suggesting that T1AM analogs may interact with these proteins and affect their physiological features.
In the present study, we investigated the possible beneficial effects of T1AM and its analogs on TTR. TTR is a transporter of thyroxine and a holo-retinol-binding protein. In its native state, TTR maintains a well-folded tetrameric complex, in which two hydrophobic binding pockets for T4 are constructed [13], [14]. Notably, in addition to its physiological importance, TTR has attracted enormous attention because of its intrinsic ability to aggregate and form amyloid fibrils. TTR amyloidosis (ATTR) is known to be caused by wild-type protein (often called senile systemic amyloidosis, as it mainly occurs at the age of 70 years) or by a single amino-acid substitution, such as V30M, L55P, and V122I, which often results in amyloid fibril deposition in the heart or peripheral nervous system (thus referred to as familial amyloid cardiomyopathy or polyneuropathy) [15], [16], [17], [18]. Previous in vitro and in vivo studies have shown that the amyloidogenic propensity of TTR correlates with its tetramer stability; dissociation into monomers and the subsequent formation of misfolded species is a rate-limiting step of TTR aggregation and fibril formation [17], [19]. Based on these observations, it was reasoned that molecules that can bind to the hydrophobic T4 binding pocket may stabilize the tetrameric state of TTR and inhibit its aggregation [20]. This idea was indeed followed by the successful development of the drug tafamidis, whose inhibitory effect on ATTR was validated in clinical trials [21], [22].
However, even after this great success, the need to develop further optimized therapeutic molecules for ATTR still exists because of the occasional limited efficacy and reduced capability of tafamidis to be distributed in the brain or eyes [23], [24]. Therefore, in the present study, we aimed to assess whether T1AM and its analogs, SG2, SG6, and SG12 (Fig. 1), interact with TTR and exert an additional role as a suppressor of ATTR. These T1AM analogs were selected based on their structural resemblance and functional efficacy, while pertaining differences in their functional groups. This series of ligands is therefore suitable to investigate the role of specific chemical moieties in interacting with TTR and suppressing its aggregation. To this end, we combined nuclear magnetic resonance (NMR) spectroscopy and computational simulation approaches to investigate the interactions between TTR and T1AM analogs at atomic resolution. We also validated the efficacy of T1AM analogs as stabilizers of the TTR tetramer with in vitro thioflavin T (ThT) fluorescence and turbidity measurements. Our results indicate that T1AM analogs are promising stabilizers of the TTR tetramer. In particular, considering their multi-functionality toward metabolic and neurodegenerative disorders, T1AM and its analogs may provide a novel and powerful therapeutic strategy for various related diseases.
Fig. 1.
Structures of endogenous 3-iodothyronamine (T1AM) and synthetic thyronamine-like analogs, SG2, SG6, and SG12.
2. Materials and methods
2.1. Drugs
T1AM and tafamidis were purchased from Sigma-Aldrich (St. Louis, MO, USA) and Cayman Chemical (Ann Arbor, MI, USA), respectively, and the thyronamine-like compounds, SG2, SG6, and SG12, were kindly provided by Prof. Rapposelli [5], [10]. Aliquots were stored at − 20 °C in DMSO as a 200 mM stock solution and diluted to the desired final concentration in the assay media.
2.2. Protein sample preparation
Human wild-type (WT), V30M, and L55P TTR proteins were recombinantly expressed in Escherichia coli and purified using previously described procedures [25]. Briefly, the pQE30 vector (Qiagen, Hilden, Germany) and E. coli M15(pREP4) competent cells were used for protein expression. To complement the relatively large size (∼55 kDa) of tetrameric WT TTR complex for NMR measurements, we employed the fractional deuteration procedure, where M9 minimal media made with 99 % deuterium oxide and supplemented with 3 g/L [U-13C]-D-glucose and 0.5 g/L 15NH4Cl (Cambridge Isotope Laboratories, Tewksbury, MA, USA) was used for protein production. Subsequently, the purification procedures were composed of sonication and centrifugation for cell debris removal, anion exchange chromatography (HiTrap Q HP; Cytiva, Marlborough, MA, USA), and the final size-exclusion chromatography (HiLoad 16/600 Superdex 75 pg; Cytiva); the latter two chromatographic procedures were conducted with the ÄKTA FPLC system (Cytiva). The fractions from the chromatographic runs were monitored using SDS-PAGE for purity. The final pure fractions were pooled, flash-frozen with liquid nitrogen, and stored in a − 80 °C freezer until use. We confirmed the integrity of the final TTR samples with mass-spectrometry and NMR; the mass-spectrometric analysis showed no indication of modification, and the NMR spectra obtained with our samples fit well with the deposited data in BMRB (see below).
2.3. NMR spectroscopy
For NMR experiments, we used an Avance III HD 850 MHz NMR spectrometer equipped with a cryogenic HCN probe (Bruker, Billerica, MA, USA). Fractionally deuterated and uniformly 13C- and 15N-labeled ([frac-2H; U-13C; U-15N]) WT-TTR were prepared at a concentration of 200 μM in a buffer consisting of 50 mM MES pH 6.5, 100 mM NaCl, 5 mM dithiothreitol, 0.01 % NaN3, and 7 % D2O. The sample volume was adjusted to 300 μL to be accommodated in a 5-mm Shigemi tube (Sigma-Aldrich). Resonance assignment information was obtained from BMRB accession numbers 5507 and 27514 [26], [27]. For NMR data acquisition, the TopSpin 3.2 software package (Bruker) was used, and we employed the POKY software suite for NMR data analysis [28].
Titration experiments were conducted by serially adding T1AM analogs (dissolved in DMSO-d6) to the [frac-2H; U-13C; U-15N]-WT-TTR sample and collecting 2D 1H–15N TROSY-HSQC spectra. We repeated the same titration experiments with tafamidis (dissolved in DMSO-d6) for comparison with those of T1AM analogs. To clarify the signal assignment of the 1:1 mixture of TTR and the T1AM analog, 3D TROSY-HNCA spectra were also obtained. Notably, we confirmed that the NMR spectra did not exhibit any noticeable signal perturbation upon the addition of the same amount of DMSO-d6 (without T1AM analogs) to the TTR sample. The signal perturbation plot was obtained by calculating ΔδNH for each signal upon addition of 1-fold T1AM analogs using the following equation: ΔδNH = [(ΔδN/5)2 + (ΔδH)2]1/2, where ΔδN and ΔδH are the signal movements in ppm in the NMR spectra of ligand-free and bound WT-TTR.
Finally, to test whether tafamidis competes with SG2 or SG6 for the same binding site of TTR, we added 1-fold tafamidis to the samples of [frac-2H; U-13C; U-15N]-WT-TTR containing 2-fold SG2 or SG6, and collected 2D 1H–15N TROSY-HSQC spectra.
2.4. Aggregation assay
The aggregation propensity of TTR samples (WT, V30M, and L55) in the absence or presence of T1AM analogs was evaluated using an aggregation assay under mildly acidic conditions [29], [30]. TTR samples, which were prepared at a concentration of 64 μM in PBS buffer (pH 7.4; 10 mM phosphate, 140 mM NaCl, and 2.7 mM KCl), were first treated with either DMSO or five-fold T1AM analogs (T1AM, SG2, SG6, or SG12) dissolved in DMSO. These TTR-T1AM analog mixtures were subsequently mixed in a 1:1 ratio with acetate buffer (200 mM sodium acetate pH 4.2, 100 mM KCl, and 1 mM EDTA). The mixture was then incubated at 37 °C without agitation.
Aggregation was monitored by ThT fluorescence (and turbidity measurements for WT TTR) using a Tecan Spark™ 10 M microplate reader (Männedorf, Switzerland). To measure ThT fluorescence, the samples were first diluted to a final concentration of 4 μM TTR with a buffer consisting of 200 mM Tris (pH 8.0) and 150 mM NaCl, and 400 μL of this mixture was then mixed with 2 μL of 2 mM ThT solution in a buffer consisting of 200 mM Tris (pH 8.0) and 150 mM NaCl. The measurements were performed in triplicate with the samples filled in a 96-well black-wall microplate (excitation and emission wavelengths were 440 nm and 482 nm, respectively). To measure turbidity, the sample was transferred into a 96-well transparent microplate, and the optical density at 330 nm was obtained in triplicate. The measurement results were averaged and plotted in the figures along with their standard deviation as an error bar. The reproducibility of the measurement results was confirmed by repeating the same experiment two to three times.
2.5. Molecular docking and molecular dynamics (MD) simulation
Protein preparation and molecular docking were performed in the TTR-T4 complex of the Protein Data Bank (PDB) [31] coded with PDB ID 2ROX [14] using the Chemscore function of the GOLD program, as previously validated [32]. MD simulations were performed to test the stability of the predicted pose using AMBER Version 16 (San Francisco, CA, USA) [33]. The TTR complexes, derived from the best scored docking poses of SG2, SG6, SG12, T1AM, and tafamidis as a control were placed in a parallelepiped water box using an explicit solvent model (TIP3P) and solvated with a 10 Å water cap. For system neutralization, sodium ions were used as counterions. Three steps of minimization were carried out before 70 ns of MD simulation: optimization of the solvent, relaxation of the side chains of the protein and finally of the ligand. We used the particle mesh Ewald electrostatic and periodic boundary conditions. For MD trajectory, minimized structures as the starting conformations were used. We set the time step of the simulations to 2.0 fs with a cutoff of 10 Å for the non-bonded interaction, and employed SHAKE to keep all bonds involving hydrogen atoms rigid. Constant-volume periodic boundary MD was run for 500 ps along with the temperature increase from 0 to 300 K. Subsequently, constant-pressure periodic boundary MD was run for 69.5 ns at 300 K. We used the Langevin thermostat to keep the temperature of the system constant, sustaining all the α carbons and ligand with 10 kcal of constant harmonic force for the first 1.6 ns. After this, we relaxed the ligand for additional 1.6 ns, and ran the last simulation without any constraint for the remaining 66.3 ns. The trajectory was collected in one frame per every 100 ps of the simulation, generating 700 frames for all complexes. General Amber force field parameters were assigned to the ligands, and partial charges were calculated using the AM1-BCC method. The set of the 30 closest water molecules to the ligand was determined using the closest command in the CPPTRAJ module [34] of AMBER 16. The solvent molecules were set automatically by distance, and the water closest for each frame was recorded. The MD trajectories were examined with the MD Movie tool of Chimera [35] and the CPPTRAJ module. The Molecular Mechanics Poisson-Boltzmann Surface Area (MMPBSA) approach [36] was used to calculate binding free energy of the complexes, applying the MMPBSA routine [37]of AMBER to 100 frames extracted from stable simulation trajectories. The contribution of each binding site residue to ligand stabilization in the same trajectory was also analyzed, using the PAIRWISE command in the CPPTRAJ module, which writes out van der Waals energy (EVDW) and electrostatic energy (EELEC) of non-bonded interactions. Results are reported as the sum of EVDW and EELEC in kcal/mol.
3. Results
3.1. NMR titration experiments of TTR with T1AM analogs
We first examined whether T1AM analogs directly interact with TTR. To this end, we collected a series of 2D 1H–15N TROSY-HSQC and 3D TROSY-HNCA spectra of the [frac-2H; U-13C; U-15N]-TTR samples titrated with increasing amounts of ligands; that is, T1AM, SG2, SG6, and SG12 (Fig. 1). Notably, all the ligands incurred NMR signal perturbation, albeit in a different fashion (Fig. 2 and S1). Upon addition of 1-equivalent of T1AM to TTR, signals corresponding to the residues V14, V20, G22, and A108 disappeared beyond detection, whereas signals for M13, K15, L17, A19, I26, S50, G53, L55-G57, T59, I107, A109-L111, S117, and V122 shifted significantly (ΔδNH > 0.02 ppm; the average plus one standard deviation value from the results of SG6 titration). SG2 induced consistent, yet more pronounced signal perturbation: the signals for A19, V20, G22, E92, A108, L110-S112, Y114, S115, S117, T118, A120, and V121 disappeared with 1-fold addition of SG2, and the signals for C10, V14-L17, S23, A25-N27, F33, S50, E51, G53, L55, L58, K76, K80, H88, E89, A91, V94, Y105-I107, A109, V122, and K126 shifted significantly. The 1-fold addition of SG6 resulted in the disappearance of the signals for K15, L17, V20, G22, A25, A108-L111, Y114, S115, S117, T118, A120, and V121, whereas the signals for V16, A19, S23, S50, L55, L58, K76, F87, E92-V94, T96, T106, I107, S112, and V122 were significantly shifted. In contrast, the overall signal perturbation was greatly diminished in the titration results of SG-12; the 1-fold addition to TTR only caused minimal signal shifts without any disappearance. We also observed consistent changes in the signal intensity upon ligand titration; as a representative example, the signal intensity change of the residue A109 is shown in Fig. S2.
Fig. 2.
Results of NMR signal perturbation induced by titrations of T1AM analogs onto TTR. One-fold equivalent (vs. the concentration of TTR monomer) of T1AM and its analogs were added to the sample of [frac-2H; U-13C; U-15N]-TTR, and the 2D 1H–15N HSQC and 3D HNCA spectra were obtained to monitor the signal perturbations of TTR. (A-D) Perturbation of the backbone 1H–15N HSQC signals by the titration of T1AM analogs was plotted using the following equation: ΔδNH (ppm) = [(ΔδN/5)2 + (ΔδH)2]1/2 (see Methods for details). The residues whose signals disappeared beyond detection during titration are indicated by red triangles. (E-G) Structural model of TTR (PDB ID: 2ROX) colored according to the extent of signal perturbations. Residues whose signals were significantly perturbed are colored as follows: red, signals that disappeared during titration; blue, signals that exhibited significant shifts (ΔδNH > 0.02 ppm); black, signals not assigned residues and prolines. On the left is the native tetrameric model, while only half of the tetrameric model is shown on the right for better presentation. The colors are shown at one subunit for simplicity. Note that the same figure was not included for the result of SG12 due to the lack of noticeable signal perturbation.
To compare the ligand binding of T1AM and its analogs, we examined whether each added ligand dissociates from TTR using a simple buffer exchange procedure. By comparing the 2D 1H–15N TROSY-HSQC spectra before and after buffer exchange, we could easily distinguish the ligand-free state of TTR from the ligand-bound state. From these trials, we confirmed that T1AM and SG12 could be easily removed from the TTR sample, whereas extensive buffer exchange could not dissociate the SG2- and SG6-TTR complexes (Fig. S3). This result indicated that the binding interactions of TTR with SG2 and SG6 were sufficiently stable, whereas those of T1AM and SG12 were not.
Overall, ligand titration experiments showed that the addition of SG2 and SG6 resulted in the most severe signal perturbations (Fig. 2). Notably, SG6 exhibited more signal broadening than SG2, while signal movements were more noticeable in the SG2 titration than in SG6. In contrast, T1AM or SG12 caused only modest or minimal signal changes, respectively.
Moreover, we tested whether tafamidis competes with T1AM analogs for the binding site on TTR. The 2D 1H–15N TROSY-HSQC spectra of the TTR samples, which mixed first with either 2-fold SG2 or SG6, and subsequently with 1-fold tafamidis, exhibited the same spectral feature with that of the TTR sample containing 1-fold tafamidis (Fig. S4). This indicates that the affinity of tafamidis for TTR was stronger than those of SG2 and SG6, and T1AM and its analogs bind to TTR with the overlapped binding site of tafamidis.
3.2. Aggregation assay of TTR in the presence of T1AM analogs
Next, we tested the efficacy of T1AM analogs in inhibiting the aggregation of TTR. The aggregation of TTR was induced by lowering the pH of the protein sample to 4.4, and the suppressive effects of T1AM analogs were tested by inducing TTR aggregation in the absence or presence of 5-fold ligands [29], [38]. Subsequent aggregation was monitored using ThT fluorescence and turbidity measurements (Fig. 3 and S5). As a reference molecule, we examined the effect of tafamidis, and we only focused on the data obtained with 5-fold ligands, because the results obtained in the presence of 1-fold T1AM analogs showed less distinguishable effects on TTR aggregation (Fig. S6). The measurement results indicated that among T1AM analogs, SG6 was the most effective inhibitor of TTR aggregation, while the effect of SG12 was negligible in ThT assays. This observation is consistent with the NMR results, where the binding interaction between TTR and SG6 was stronger than the others, whereas SG12 caused minimal perturbations to the NMR signals of TTR. In contrast, the relatively weak inhibitory effects of SG2 on TTR aggregation were somewhat inconsistent with the NMR results because the NMR signal perturbations by SG2 were comparable with those by SG6.
Fig. 3.
TTR aggregation assay results in the presence/absence of 5-fold T1AM and its analogs. The results without any ligand (DMSO) or with 5-fold tafamidis (Tafa) were included for comparison. TTR aggregation was induced by mildly acidic conditions (the concentration of TTR was 32 μM) and was subsequently quantified using thioflavin-T (ThT) fluorescence (A) and turbidity (B) measurements after 3-day incubation at 37 °C (the daily measurement results of ThT fluorescence are shown in Fig. S5). The measurements were conducted in triplicate, and the standard deviations from those were denoted as error bars in the figure.
In addition, we repeated the same aggregation assay with two pathogenic variants of TTR, V30M and L55P, which exhibited more amyloidogenic propensities than WT [39], [40]. Upon treatment of 5-fold T1AM analogs to V30M TTR, TTR aggregation was again slowed down significantly; the inhibitory effect of SG6 was the most pronounced among T1AM analogs (Fig. 4A and S7A). In contrast, T1AM analogs could not cause any noticeable effect on the aggregation of L55P TTR (Fig. 4B and S7B).
Fig. 4.
Aggregation assay results of two pathogenic TTR variants, V30M (A) and L55P (B), in the presence/absence of 5-fold T1AM and its analogs. The results without any ligand (DMSO) or with 5-fold tafamidis (Tafa) were included for comparison. TTR aggregation was induced by mildly acidic conditions (the concentration of TTR was 32 μM) and was subsequently quantified using thioflavin-T (ThT) fluorescence measurements after 3-day incubation at 37 °C (the daily measurement results of ThT fluorescence are shown in Fig. S7). The measurements were conducted in triplicate, and the standard deviations from those were denoted as error bars in the figure.
3.3. Molecular docking simulation of TTR with T1AM analogs
To investigate the details of the interaction between TTR and T1AM analogs at atomic resolution, we conducted molecular docking simulations and compared the resultant complex models. The binding of T4 with TTR includes the ionic bridge between K15 (TTR) and the carboxylic acid (T4) and the polar interaction between E54 (TTR) and the protonated amine (T4) [14]. Therefore, we aimed to investigate the interaction of the single amine (for T1AM, SG2, and SG12) or carboxylate (for SG6) moiety of T1AM analogs in the TTR binding site using molecular docking simulation, comparing results to the pose of tafamidis (Fig. 5) [21].
Fig. 5.
Superposition of the docking results (gray), and the average structure of MD simulation of SG2 (A; pink), SG12 (B; tun), T1AM (C; green), SG6 (D; cyan) and tafamidis (E; orange). The chain (A, B, C, or D) from which each residue originates is indicated along with the residue information.
Overall, the docking models provided a detailed view of the interactions between TTR and T1AM analogs (Fig. 5, gray). First, although the docking model predicted that both SG2 and SG12 interact similarly in the TTR-binding site, the longer spacer of SG2 allows a stronger interaction with E54 of TTR (Fig. 5AB, gray). The aniline moiety of SG2 and SG12 lies in the inner region of TTR, towards S115, S117, and T119. In contrast, the molecular docking results for T1AM were completely different; a reversed pose of T1AM was suggested, thus placing the phenolic OH group near K15 (Fig. 5C, gray). This interaction is common and frequent within ligands crystallized with TTR [41], [42] and is preferred over the unusual interaction between the amine and E54. The amine chain occupies the serine-rich region, engaging S117 of both chains A and C, whereas the same locus was accompanied by the aniline group of SG compounds. Docking of SG6 (Fig. 5D, gray) is, as expected, in the classical forward mode, guaranteeing the ionic bond between carboxylate and K15 and the polar interaction between the aniline moiety and S117. The binding mode described here for T1AM analogs is very similar to the classical arrangement of other kinetic stabilizers of TTR, such as tafamidis (Fig. 5E, gray) [21]: a strong ionic binding anchors the carboxylate to K15, in the outer region of the binding site, and many lipophilic interactions stabilize the ligand in the central part of the pocket, which is composed by three parallel sheets (residues 15–17, 108–110 and 117–119 for both specular chains of interfaced dimers). The two chlorines protrude towards the inner polar pocket, rich in serines (S115 and S117) belonging to all neighboring monomers, and strongly involved in the stabilization of the TTR tetramer [43].
3.4. Molecular dynamics (MD) simulation of the TTR-ligand complexes
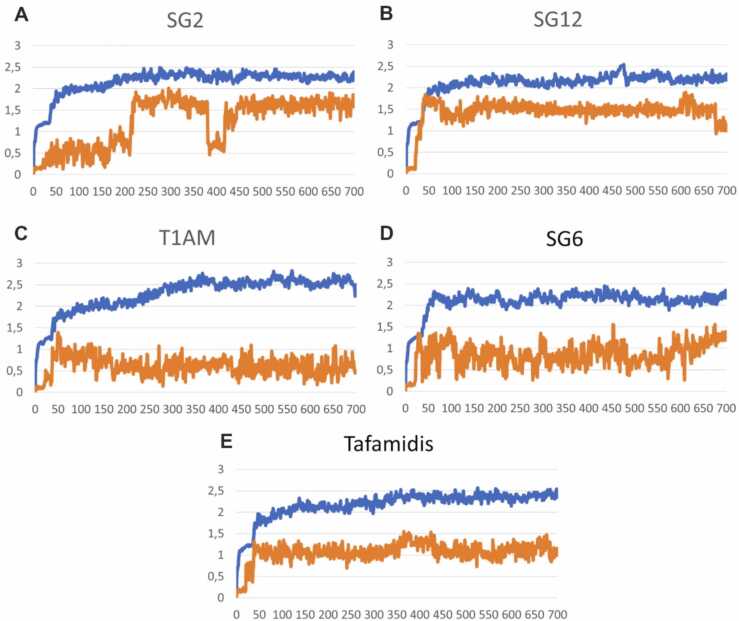
To further evaluate the stability of the complexes between TTR and T1AM analogs, we conducted MD simulations. After constraint relaxation (frame 40), the protein backbone reached a Cα RMSD stability of 2 Å in all complexes (Fig. 6). In the TTR-SG2 complex, (Fig. 6A) a shift of the RMSD plot is detected, due to the specular movement of the ligand between the two interfaced dimers, near frame 400; the RMSD of SG2 returned stable at an average value of 1.5 Å, like all the other ligands.
Fig. 6.
RMSD of the TTR Cα atoms (blue line) and ligand heavy atoms (orange line) in molecular dynamics simulations of the TTR-ligand complexes: SG2-TTR (A), SG12-TTR (B), T1AM-TTR (C), SG6-TTR (D) and tafamidis-TTR (E).
Ultimately, despite some system fluctuations, all compounds retained their binding disposition. The final structures of the MD simulations superimposed on the initial docking pose are shown in Fig. 5. It was evident from the model of SG12-TTR complex (Fig. 5B, tan) that K15 attempted to reach E54 and detach it from the protonated amine of the ligand. Not only the initial interaction with E54 failed, the interactions with serines in the inner pocket were also lost. The behavior of SG2 in complex with TTR was different (Fig. 5A, pink): although the initial interaction between the aminoethyloxy chains of SG2 and E54 was retained during the simulation, it failed to maintain stable interaction with S117 and T119, probably because the molecule length is not sufficient to reach the inner region when the protonated amine anchors E54. T1AM (Fig. 5C, green), in its inverted pose, maintained a polar interaction with K15, which appeared weaker than a hydrogen bond, because K15 was engaged in strong ionic bonding with E54. The ligand still seemed to engage in fruitful interactions with serine residues. The comparison with SG6 (Fig. 5D, cyan), showed how the usual ionic interaction with K15 of the carboxylate group of SG6 guarantees a better interaction with respect to the similar docking pose of SG2. Fig. 5E shows the results obtained with tafamidis, which were used as a control and subjected to the same docking and dynamics procedure. Notably, the final structure of tafamidis overlaps well with the experimental pose, validating the suitability of our approaches.
To explore the role of the key residues in ligand stabilization and their correlation with inhibitory activity against TTR aggregation, we analyzed the MD trajectories in detail (Fig. 7). This helped us to identify the effect of fluctuations on interactions between the residues and the ligands over time, which were not discernible in Fig. 5. Residues involved in the interaction with a distance less than 5 Å from the ligand were analyzed, with a focus on two regions: the outer region, which usually binds with K15, and the inner region, which interacts with S115 and S117. Water molecules were also analyzed separately: the one closest (less than 3 Å apart) to the ligand end in the K15 region, and the other closest to the opposite end of the ligand in the inner region.
Fig. 7.
Distances of the key residues of TTR from the ligand during molecular dynamics simulation of the TTR-ligand complexes. Only residues involved within distances less than 5 Å are reported: K15 (magenta), E54 (red), S115D (S115 from the chain D; green), S117A (gray), S117C (black), T119C (gold). For K15 and E54, due to the high flexibility, the closest value between chains A and C has been reported for clarity. Water molecules less than 3 Å from the protonated amine of SG2 and SG12, the phenolic hydroxyl of T1AM, and the carboxylate of SG6 and tafamidis are reported in blue (WAT OUT), whereas water molecules less than 3 Å from the aniline nitrogen of SG2, SG12 and SG6 and chlorines of tafamidis are reported in cyan (WAT IN).
For SG2 (Fig. 7A), the involvement of many residues, particularly S117 and T119, is evident in more than half of the unconstrained simulation. In addition, E54 stably interact with the amine of SG2. The protonated amine group of K15 was located approximately 4.5 Å from the amine group of SG2 for the entire simulation, interacting with E54 and weakly with the ether oxygen atom of the SG2 linker (not reported in Fig. 5 for clarity). The first level of interactions is however with water, in both the outer and inner regions of the binding sites. This means that during the simulation the interaction of SG2 with E54, S117 and T119 was recurrently displaced by the solvent. Regarding SG12 (Fig. 7B), the initial interaction between SG12 and E54 loosened during the unconstrained simulation, as did the nearness of K15. Only fluctuating interactions between the aniline group and residues S117 and T119 (Fig. 7B; black and orange lines, respectively) were visible. In a large part of the simulation, the main interaction in the inner and outer regions was with water molecules, which was consistent with its relatively weak interaction with TTR. T1AM engaged in fluctuating interactions with K15 by the phenolic OH group (Fig. 7C; magenta line) and a polar interaction between the protonated amine of T1AM and S117 with an average distance of approximately 4 Å (Fig. 7C; gray, distance between heteroatoms), and with T119 (Fig. 7C; orange, distance between heteroatoms). However, in the inner region, the main direct interaction was with water molecules; the distance between the protonated amine of T1AM and the closest water molecules during the simulation was reported to be less than 3 Å (Fig. 7C; cyan). A large number of water molecules seemed to be also attracted in the outer region of the binding site (Fig. 7C; blue). For SG6, strong interactions with S117 and T119 were evident without any water molecule interference or bridges (Fig. 7D). During the simulation a strong interaction was observed between the carboxylate group of SG6 and K15 (alternatively K15 of the A and C chains: the minimum distance is shown in the figure). The interactions with K15, S115, S117, and T119 were maintained, displacing the water that is poorly represented in the binding site. The average number of water molecules per frame of simulation in the region of K15 varies from about 2 for SG2 and SG12, to 1 for T1AM and 0.02 for SG6 (0.03 for tafamidis; see Table 1). Intriguingly, high hydration was detected in the inner region for T1AM, due to its reversed pose placing the protonated amine in this pocket, while no water molecules for tafamidis.
Table 1.
Mean number of water molecules per molecular dynamics frame less than 3 Å from the protonated amine of SG2 and SG12, the phenolic hydroxyl of T1AM, and the carboxylate of SG6 and tafamidis (WAT OUT), and from the aniline nitrogen of SG2, SG12, and SG6 and chlorines of tafamidis (WAT IN).
| WAT OUT | WAT IN | |
|---|---|---|
| SG2 | 2 | 0.5 |
| SG12 | 2.5 | 0.5 |
| T1AM | 1.2 | 2 |
| SG6 | 0.02 | 0.1 |
| Tafamidis | 0.03 | 0 |
With the aim to validate the binding affinity information suggested by molecular dynamics simulation, a MMPBSA calculation was performed on all complexes (Table 2). In this calculation, T1AM and SG12 complexes showed unfavorable binding energy, and SG2 showed slightly negative energy. On the other hand, SG6 and tafamidis have a favorable free energy, which is in agreement with our experimental results.
Table 2.
Binding free energy of TTR complexes, obtained by the MMPBSA method. VDWAALS = van der Waals contribution; EEL = electrostatic energy; EPB = electrostatic contribution to the solvation free energy calculated by Poisson-Boltzmann method; ENPOLAR = nonpolar solvation free energy contribution; EDISPER = dispersion term; ΔGtot = final estimated binding free energy in kcal/mol.
| ENERGY (kcal/mol) | SG2 | SG12 | T1AM | SG6 | Tafamidis |
|---|---|---|---|---|---|
| EEL | -28.18 | -29.78 | -38.44 | -30.65 | -36.10 |
| WDV | -236.43 | -212.91 | -241.22 | 107.02 | 124.46 |
| EPB | 246.31 | 228.47 | 260.06 | -105.57 | -116.50 |
| ENPOLAR | -24.44 | -23.53 | -26.68 | -25.74 | -25.71 |
| EDISPER | 40.77 | 39.63 | 46.96 | 43.23 | 45.94 |
| ΔGtot | -1.97 | 1.87 | 0.67 | -11.72 | -7.90 |
Taken together, the results of the MD simulations provide placeable explanation to rationalize the results of the NMR titration experiments and the TTR aggregation assay. An additional effort was made to evaluate the energetic correlation between the NMR data and the closest atoms along the MD trajectory, through a per-residue deconvolution of the interaction energies. The pairwise calculation of the interaction energy between ligands and each binding-site residue is summarized in Fig. 8. No residue in the binding pocket of the SG12-TTR complex was involved in strong interaction with the ligand, except E54. In the T1AM-TTR complex, many residues are involved in weak interactions, covering the central region of the binding site (L17, A108, A109, and L110). Despite the negative term of the electrostatic energy between T1AM and S115, S117, and T119, an unfavorable VDW component led to a slightly positive contribution for the serines, more marked for T119. SG2 seemed, differently, to strongly interact with the central part of the binding site (L17, A108, A109, and L110), and with E54. The latter result was overestimated because the pairwise calculation cannot account for the presence of water, which is crucial for the ionic interaction between E54 and the protonated amine of SG2. Negative values of interaction energy were also detected for the polar residues of the inner region, in agreement with the NMR perturbation results. K15 is involved only in the binding of SG6, with high negative values of energy. For the SG6-TTR complex, the interaction spectrum of the residues is similar to that of SG2, yet with stronger energies. Negative values were detected for L17, A108, A109, L110, S115, S117, and T119.
Fig. 8.
Results of the pairwise non-bonded energy calculations. The contribution of each residue of the TTR binding site to the ligand stabilization was reported in terms of kcal/mol of interaction energy with the color code labeled as follows: E > 16 kcal/mol colored in black; 0 < E < 16 kcal/mol in dark gray; − 10 < E < 0 kcal/mol in light gray; − 30 < E < −10 kcal/mol in pink; E < −30 kcal/mol in red.
Notably, molecular dynamics results and energy calculations demonstrate that the presence of a protonated amine, potentially capable to bind E54 analogously to T4, leads to attracting water in the binding site [44]. SG12 lost its initial interaction with S117 and T119; SG2, with the aminoethoxy linker, could engage a strong ionic bond with E54 and retain polar interaction with S115, S117 and T119, but they were substituted by water molecules during the simulation, perhaps due to insufficient chain length. T1AM, despite the high flexibility of the amine chain and its polarity, which could induce a strong interaction with serine residues and T119, attracts stable water molecules in the range of 3 Å from the amine, losing direct contact with the binding site residues.
4. Discussion
The distinct features of thyromimetic molecules in various pathological processes have attracted a wide range of interest. However, despite numerous studies evaluating various beneficial effects of thyromimetics, their interaction with TTR, the TH transporter protein, and the subsequent effects on several TTR-related pathogenic mechanisms, for example, ATTR, have not been sufficiently investigated [45]. To this end, we previously characterized the interaction between TTR and diphenyl-methane-based thyromimetics (sobetirome and its analogs, IS25 and TG68) using NMR spectroscopy and computational simulation, and reported that these molecules interact with and effectively suppress TTR aggregation [46]. In the present study, we aimed to elucidate the binding interaction between TTR and thyronamine-based compounds, including 3-iodothyronamine (T1AM) and its diphenyl-methane analogs (i.e., SG2, SG6, and SG12) and characterize their subsequent effects on TTR aggregation. Although a series of T1AM analogs have been noted as promising therapeutic molecules for obesity and NDDs [11], [47], their relationship with TTR has not been elucidated to date.
Our NMR spectroscopic titration experiments confirmed the direct binding of T1AM and its analogs at the hydrophobic T4-binding site of TTR (Fig. 2 and S4). However, subsequent NMR signal perturbation analyses indicated that each T1AM analog exerted differential effects on TTR NMR signals. First, it was evident that the TTR signals were most affected by SG2 and SG6, whereas the effects of T1AM and SG12 were less significant. Moreover, in contrast to the wide signal perturbations by SG2 and SG6 at both the inner and outer regions of the T4-binding pocket, the NMR signal perturbation by T1AM was localized at the outer region, whereas that by SG12 was more focused at the inner region of the binding pocket (Fig. 2). ThT fluorescence and turbidity assay results further corroborated the NMR data (Fig. 3). The activity of T1AM analogs in suppressing acid-induced aggregation of WT TTR was high in the following order: SG6 > SG2 ≈ T1AM > SG12. This correlates well with the NMR signal perturbation (Fig. 2) and intensity change (Fig. S2) results; more significant changes in TTR NMR signals imply a stronger interaction, which subsequently suggests higher stability of the native tetrameric state of TTR.
Intriguingly, the aggregation assays of two pathogenic variants of TTR, V30M and L55P, indicated that the efficacy of T1AM analogs manifests with WT and V30M, not with L55P. This may be attributed to the altered T4 binding site of L55P TTR. Based on our calculation results, E54 play critical roles to stabilize the interaction with T1AM and its analogs. The V30M substitution may cause only, if any, a minimal effect on the binding site architecture, while the effect of L55P substitution would be more significant due to its proximal location to E54. Indeed, the structural model of L55P TTR indicated that the conformation of E54 can be disturbed by the mutation [48]. These observations collectively suggest that a certain TTR variant may require additional consideration for ligand design to ensure its sufficient functionality toward TTR aggregation.
Molecular docking calculations and MD simulation results also provided consistent, yet more detailed explanations for the differential binding interactions of T1AM analogs with TTR. First, the simulation results consistently indicated that SG2 and SG6 were tighter binders of TTR than T1AM and SG12. The intriguing difference in the NMR titration results of SG2 and SG6, however, is that the region including the residues S50-T60 was more affected by the interaction with SG2 than that with SG6. The hints for this came from the simulations suggesting the aminoethoxy chain of SG2 may mediate the interaction with E54, while the carboxylate group of SG6 directly interacts with K15. It is also notable that SG6 exhibited the most inhibitory effects on TTR aggregation, implying the importance of the local interaction between the carboxylate group of the ligand and the amine group of K15 for an aggregation suppressor; a similar observation was made in previous reports that some thyromimetic suppressors of TTR aggregation, such as tafamidis and GC-1 analogs, also contain the terminal carboxylate group that may be involved in the interaction with K15 [21], [46]. The simulation results also provide plausible explanations for the NMR signal perturbation results of T1AM and SG12. For T1AM, the computational study predicted the opposite binding and incorporation of water molecules in the region of the T4 binding pocket, resulting in the formation of a complex with reduced stability. The calculations showed SG12 to be a weak binder, mainly due to the lack of a stable interaction network with K15 and E54, in addition to the strong hydration of the amine group. These observations suggest that the single amine moiety of T1AM analogs, in contrast to the single carboxylate, could be critical for the interaction with TTR; they need to have an appropriate linker accommodating the network with K15 and E54, as well as hydrophobic and bulky moieties complementing the inner hydrophobic T4 binding pocket of TTR. Finally, the MMPBSA calculation results summarized these analyses to conclude that the binding affinity of T1AM analogs for TTR is in the following order: SG6 > SG2 > T1AM > SG12, which aligns well with the NMR data and the aggregation assay results.
Taken together, our study provides a novel possibility for using T1AM and its analogs as therapeutic molecules to modulate TTR amyloidosis. Owing to the well-established benefits of TH-derived metabolites and thyromimetics in various human diseases, many related derivatives and analogs have been developed and investigated [2], [11], [49], [50]. In this study, although it was clear that T1AM and its analogs are not as efficient as tafamidis for suppressing the aggregation of TTR, we could still demonstrate that T1AM and its analogs, SG2 and SG6, have therapeutic potentials toward TTR aggregation and prove the feasibility of developing further optimized multi-functional molecules that are effective for both TTR amyloidosis and TH-related metabolic disorders or neurodegeneration.
To the best of our knowledge, this is the first study to observe the direct binding of T1AM and thyronamine-like analogs to TTR. Although a previous study showed that the major transporter of T1AM is apolipoprotein B-100 [51], [52], our observations provide a plausible clue that TTR may work as an additional transporter protein at least for some T1AM analogs. This indicates that T1AM analogs may have multiple transport pathways, resulting in differential kinetic profiles and distinctive distributions. Therefore, we believe that the present study may provide novel insights into the working mechanisms of T1AM analogs and how to maximize their efficacies.
Funding
This research was supported by the National Research Foundation funded by the Ministry of Science and ICT, Republic of Korea (grant number NRF-2018R1C1B6008282 to J.H.K.) and the Institute for Basic Science, Republic of Korea (grant number IBS-R007-D1 to Y.H.K). and Pfizer (Project ID: 67562227; J.H.K. and G.C.).
CRediT authorship contribution statement
Bokyung Kim: Conceptualization, Data curation, Investigation, Methodology, Validation, Writing – original draft. Young Ho Ko: Data curation, Investigation, Resources. Jinbeom Si: Investigation. Jongbum Na: Investigation. Gabriella Ortore: Conceptualization, Data curation, Investigation, Methodology, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. Grazia Chiellini: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing. Jin Hae Kim: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Declarations of Competing Interest
None.
Acknowledgments
The authors are indebted to Professor Kimoon Kim (Center for Self-assembly and Complexity, Institute for Basic Science) for his considerate support in using the NMR spectrometers. The authors also gratefully acknowledge Prof. Simona Rapposelli (Department of Pharmacy, University of Pisa) for providing SG-2, SG-6, and SG-12 as a gift.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.09.028.
Contributor Information
Gabriella Ortore, Email: gabriella.ortore@unipi.it.
Grazia Chiellini, Email: grazia.chiellini@unipi.it.
Jin Hae Kim, Email: jinhaekim@dgist.ac.kr.
Appendix A. Supplementary material
Supplementary material.
.
References
- 1.Köhrle J., Biebermann H. 3-Iodothyronamine-A thyroid hormone metabolite with distinct target profiles and mode of action. Endocr Rev. 2019;40:620–630. doi: 10.1210/er.2018-00182. [DOI] [PubMed] [Google Scholar]
- 2.Gauthier B.R., Sola‐García A., Cáliz‐Molina M.Á., Lorenzo P.I., Cobo‐Vuilleumier N., Capilla‐González V., et al. Thyroid hormones in diabetes, cancer, and aging. Aging Cell. 2020;19:1–25. doi: 10.1111/acel.13260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scanlan T.S., Suchland K.L., Hart M.E., Chiellini G., Huang Y., Kruzich P.J., et al. 3-Iodothyronamine is an endogenous and rapid-acting derivative of thyroid hormone. Nat Med. 2004;10:638–642. doi: 10.1038/nm1051. [DOI] [PubMed] [Google Scholar]
- 4.Cöster M., Biebermann H., Schöneberg T., Stäubert C. Evolutionary conservation of 3-iodothyronamine as an agonist at the trace amine-associated receptor 1. Eur Thyroid J. 2015;4:9–20. doi: 10.1159/000430839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellusci L., Laurino A., Sabatini M., Sestito S., Lenzi P., Raimondi L., et al. New insights into the potential roles of 3-iodothyronamine (T1AM) and newly developed thyronamine-like TAAR1 agonists in neuroprotection. Front Pharm. 2017;8:1–17. doi: 10.3389/fphar.2017.00905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.di Leo N., Moscato S., Borso’ M., Sestito S., Polini B., Bandini L., et al. Delivery of thyronamines (Tams) to the brain: a preliminary study. Molecules. 2021;26:1616. doi: 10.3390/molecules26061616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bellusci L., Runfola M., Carnicelli V., Sestito S., Fulceri F., Santucci F., et al. Endogenous 3-iodothyronamine (T1Am) and synthetic thyronamine-like analog SG-2 act as novel pleiotropic neuroprotective agents through the modulation of SIRT6. Molecules. 2020;25:1–14. doi: 10.3390/molecules25051054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laurino A., Gencarelli M., Raimondi L. The 3-iodothyronamine (T1AM) and the 3-iodothyroacetic acid (TA1) indicate a novel connection with the histamine system for neuroprotection. Eur J Pharmacol. 2021;912 doi: 10.1016/j.ejphar.2021.174606. [DOI] [PubMed] [Google Scholar]
- 9.Musilli C., De Siena G., Manni M.E., Logli A., Landucci E., Zucchi R., et al. Histamine mediates behavioural and metabolic effects of 3-iodothyroacetic acid, an endogenous end product of thyroid hormone metabolism. Br J Pharmacol. 2014;171:3476–3484. doi: 10.1111/bph.12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiellini G., Nesi G., Digiacomo M., Malvasi R., Espinoza S., Sabatini M., et al. Design, synthesis, and evaluation of thyronamine analogues as novel potent mouse trace amine associated receptor 1 (m TAAR1) agonists. J Med Chem. 2015;58:5096–5107. doi: 10.1021/acs.jmedchem.5b00526. [DOI] [PubMed] [Google Scholar]
- 11.Rutigliano G., Bandini L., Sestito S., Chiellini G. 3-iodothyronamine and derivatives: new allies against metabolic syndrome? Int J Mol Sci. 2020;21 doi: 10.3390/ijms21062005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartalena L., Robbins J. Thyroid hormone transport proteins. Clin Lab Med. 1993;13:583–598. doi: 10.1016/S0272-2712(18)30427-X. [DOI] [PubMed] [Google Scholar]
- 13.Hamilton J.A., Steinrauf L.K., Braden B.C., Liepnieks J., Benson M.D., Holmgren G., et al. The X-ray crystal structure refinements of normal human transthyretin and the amyloidogenic Val-30 → Met variant to 1.7-Å resolution. J Biol Chem. 1993;268:2416–2424. doi: 10.1016/s0021-9258(18)53792-3. [DOI] [PubMed] [Google Scholar]
- 14.Wojtczak A., Cody V., Luft J.R., Pangborn W. Structures of human transthyretin complexed with thyroxine at 2.0 Å resolution and 3′,5′-dinitro- N -acetyl- L -thyronine at 2.2 Å resolution. Acta Crystallogr D Biol Crystallogr. 1996;52:758–765. doi: 10.1107/S0907444996003046. [DOI] [PubMed] [Google Scholar]
- 15.Costa P.P., Figueira A.S., Bravo F.R. Amyloid fibril protein related to prealbumin in familial amyloidotic polyneuropathy. Proc Natl Acad Sci USA. 1978;75:4499–4503. doi: 10.1073/pnas.75.9.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westermark P., Sletten K., Johansson B., Cornwell G.G. Fibril in senile systemic amyloidosis is derived from normal transthyretin. Proc Natl Acad Sci USA. 1990;87:2843–2845. doi: 10.1073/pnas.87.7.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson S.M., Connelly S., Fearns C., Powers E.T., Kelly J.W. The transthyretin amyloidoses: from delineating the molecular mechanism of aggregation linked to pathology to a regulatory-agency-approved drug. J Mol Biol. 2012;421:185–203. doi: 10.1016/j.jmb.2011.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grogan M., Scott C.G., Kyle R.A., Zeldenrust S.R., Gertz M.A., Lin G., et al. Natural history of wild-type transthyretin cardiac amyloidosis and risk stratification using a novel staging system. J Am Coll Cardiol. 2016;68:1014–1020. doi: 10.1016/j.jacc.2016.06.033. [DOI] [PubMed] [Google Scholar]
- 19.Si J.B., Kim B., Kim J.H. Transthyretin misfolding, a fatal structural pathogenesis mechanism. Int J Mol Sci. 2021;22 doi: 10.3390/ijms22094429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miroy G.J., Lai Z., Lashuel H.A., Peterson S.A., Strang C., Kelly J.W. Inhibiting transthyretin amyloid fibril formation via protein stabilization. Proc Natl Acad Sci USA. 1996;93:15051–15056. doi: 10.1073/pnas.93.26.15051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bulawa C.E., Connelly S., DeVit M., Wang L., Weigel C., Fleming J.A., et al. Tafamidis, a potent and selective transthyretin kinetic stabilizer that inhibits the amyloid cascade. Proc Natl Acad Sci USA. 2012;109:9629–9634. doi: 10.1073/pnas.1121005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maurer M.S., Schwartz J.H., Gundapaneni B., Elliott P.M., Merlini G., Waddington-Cruz M., et al. Tafamidis treatment for patients with transthyretin amyloid cardiomyopathy. N Eng J Med. 2018;379:1007–1016. doi: 10.1056/nejmoa1805689. [DOI] [PubMed] [Google Scholar]
- 23.Casal I., Monteiro S., Beirão J.M. Tafamidis in hereditary ATTR amyloidosis – our experience on monitoring the ocular manifestations. Amyloid. 2016;23:262–263. doi: 10.1080/13506129.2016.1236332. [DOI] [PubMed] [Google Scholar]
- 24.Salvi F., Volpe R., Pastorelli F., Bianchi A., Vella A., Rapezzi C., et al. Failure of tafamidis to halt progression of Ala36Pro TTR oculomeningovascular amyloidosis. J Stroke Cereb Dis. 2018;27:e212–e214. doi: 10.1016/j.jstrokecerebrovasdis.2018.04.033. [DOI] [PubMed] [Google Scholar]
- 25.Kim J.H., Oroz J., Zweckstetter M. Structure of monomeric transthyretin carrying the clinically important T119M mutation. Angew Chem Int Ed. 2016;55:16168–16171. doi: 10.1002/anie.201608516. [DOI] [PubMed] [Google Scholar]
- 26.Liu K., Kelly J.W., Wemmer D.E. Native state hydrogen exchange study of suppressor and pathogenic variants of transthyretin. J Mol Biol. 2002;320:821–832. doi: 10.1016/S0022-2836(02)00471-0. [DOI] [PubMed] [Google Scholar]
- 27.Leach B.I., Zhang X., Kelly J.W., Dyson H.J., Wright P.E. NMR measurements reveal the structural basis of transthyretin destabilization by pathogenic mutations. Biochemistry. 2018;57:4421–4430. doi: 10.1021/acs.biochem.8b00642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee W., Rahimi M., Lee Y., Chiu A. POKY: a software suite for multidimensional NMR and 3D structure calculation of biomolecules. Bioinformatics. 2021;37:3041–3042. doi: 10.1093/bioinformatics/btab180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai Z., Colón W., Kelly J.W. The acid-mediated denaturation pathway of transthyretin yields a conformational intermediate that can self-assemble into amyloid. Biochemistry. 1996;35:6470–6482. doi: 10.1021/bi952501g. [DOI] [PubMed] [Google Scholar]
- 30.Robinson L.Z., Reixach N. Quantification of quaternary structure stability in aggregation-prone proteins under physiological conditions: the transthyretin case. Biochemistry. 2014;53:6496–6510. doi: 10.1021/bi500739q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., et al. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ortore G., Martinelli A. Identification of transthyretin fibril formation inhibitors using structure-based virtual screening. ChemMedChem. 2017;12:1327–1334. doi: 10.1002/cmdc.201700051. [DOI] [PubMed] [Google Scholar]
- 33.D.A. Case, R.M. Betz, D.S. Cerutti, T.E. Cheatham, III, T.A. Darden, R.E. Duke, T.J. Giese HG, A.W. Goetz, N. Homeyer, S. Izadi, P. Janowski, J. Kaus, A. Kovalenko, T.S. Lee, S. LeGrand, P. Li C., Lin, T. Luchko, R. Luo, B. Madej, D. Mermelstein, K.M. Merz, G. Monard, H. Nguyen, H.T. Nguyen I., Omelyan, A. Onufriev, D.R. Roe, A. Roitberg, C. Sagui, C.L. Simmerling, W.M. Botello-Smith JS, R.C. Walker, J. Wang, R.M. Wolf, X. Wu LX and P.A.K.. AMBER 2016, University of California, San Francisco 2016.
- 34.Roe D.R., Cheatham T.E. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 35.Pettersen E., Goddard T., Huang C., Couch G., Greenblatt D., Meng E., et al. UCSF Chimera -- a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 36.Wang C., Nguyen P.H., Pham K., Huynh D., Le T.B.N., Wang H., et al. Calculating protein-ligand binding affinities with MMPBSA: method and error analysis. J Comput Chem. 2016;37:2436–2446. doi: 10.1002/jcc.24467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller B.R., McGee T.D., Swails J.M., Homeyer N., Gohlke H., Roitberg A.E. MMPBSA.py: An efficient program for end-state free energy calculations. J Chem Theory Comput. 2012;8:3314–3321. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- 38.Colon W., Kelly J.W. Partial denaturation of transthyretin is sufficient for amyloid fibril formation in vitro. Biochemistry. 1992;31:8654–8660. doi: 10.1021/bi00151a036. [DOI] [PubMed] [Google Scholar]
- 39.Jacobson D.R., McFarlin D.E., Kane I., Buxbaum J.N. Transthyretin Pro 55, a variant associated with early-onset, aggressive, diffuse amyloidosis with cardiac and neurologic involvement. Hum Genet. 1992;89:353–356. doi: 10.1007/BF00220559. [DOI] [PubMed] [Google Scholar]
- 40.Jacobson D.R., Buxbaum J.N. Genetic aspects of amyloidosis. Adv Hum Genet. 1991;20:69–123. doi: 10.1007/978-1-4684-5958-6_2. [DOI] [PubMed] [Google Scholar]
- 41.Cotrina E.Y., Santos L.M., Rivas J., Blasi D., Leite J.P., Liz M.A., et al. Targeting transthyretin in Alzheimer’s disease: drug discovery of small-molecule chaperones as disease-modifying drug candidates for Alzheimer’s disease. Eur J Med Chem. 2021;226 doi: 10.1016/j.ejmech.2021.113847. [DOI] [PubMed] [Google Scholar]
- 42.Mizuguchi M., Yokoyama T., Okada T., Nakagawa Y., Fujii K., Nabeshima Y., et al. Benziodarone and 6-hydroxybenziodarone are potent and selective inhibitors of transthyretin amyloidogenesis. Bioorg Med Chem. 2023;90 doi: 10.1016/j.bmc.2023.117370. [DOI] [PubMed] [Google Scholar]
- 43.Haupt M., Blakeley M.P., Fisher S.J., Mason S.A., Cooper J.B., Mitchell E.P., et al. Binding site asymmetry in human transthyretin: insights from a joint neutron and X-ray crystallographic analysis using perdeuterated protein. IUCrJ. 2014;1:429–438. doi: 10.1107/S2052252514021113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu D., Wyttenbach T., Bowers M.T. Hydration of protonated primary amines: effects of intermolecular and intramolecular hydrogen bonds. Int J Mass Spectrom. 2004;236:81–90. doi: 10.1016/J.IJMS.2004.05.013. [DOI] [Google Scholar]
- 45.Saponaro F., Kim J.H., Chiellini G. Transthyretin stabilization: an emerging strategy for the treatment of alzheimer’s disease? Int J Mol Sci. 2020;21:1–13. doi: 10.3390/ijms21228672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim B., Ko Y.H., Runfola M., Rapposelli S., Ortore G., Chiellini G., et al. Diphenyl-methane based thyromimetic inhibitors for transthyretin amyloidosis. Int J Mol Sci. 2021;22 doi: 10.3390/ijms22073488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Runfola M., Perni M., Yang X., Marchese M., Bacci A., Mero S., et al. Identification of a thyroid hormone derivative as a pleiotropic agent for the treatment of Alzheimer’s disease. Pharmaceuticals. 2021;14 doi: 10.3390/ph14121330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saelices L., Johnson L.M., Liang W.Y., Sawaya M.R., Cascio D., Ruchala P., et al. Uncovering the mechanism of aggregation of human transthyretin. J Biol Chem. 2015;290:28932–28943. doi: 10.1074/jbc.M115.659912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brenta G., Danzi S., Klein I. Potential therapeutic applications of thyroid hormone analogs. Nat Clin Pract Endocrinol Metab. 2007;3:632–640. doi: 10.1038/ncpendmet0590. [DOI] [PubMed] [Google Scholar]
- 50.Zucchi R. Thyroid hormone analogues: an update. Thyroid. 2020;30:1099–1105. doi: 10.1089/thy.2020.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roy G., Placzek E., Scanlan T.S. ApoB-100-containing lipoproteins are major carriers of 3-iodothyronamine in circulation. J Biol Chem. 2012;287:1790–1800. doi: 10.1074/jbc.M111.275552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janssen S.T., Janssen O.E. Directional thyroid hormone distribution via the blood stream to target sites. Mol Cell Endocrinol. 2017;458:16–21. doi: 10.1016/j.mce.2017.02.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.