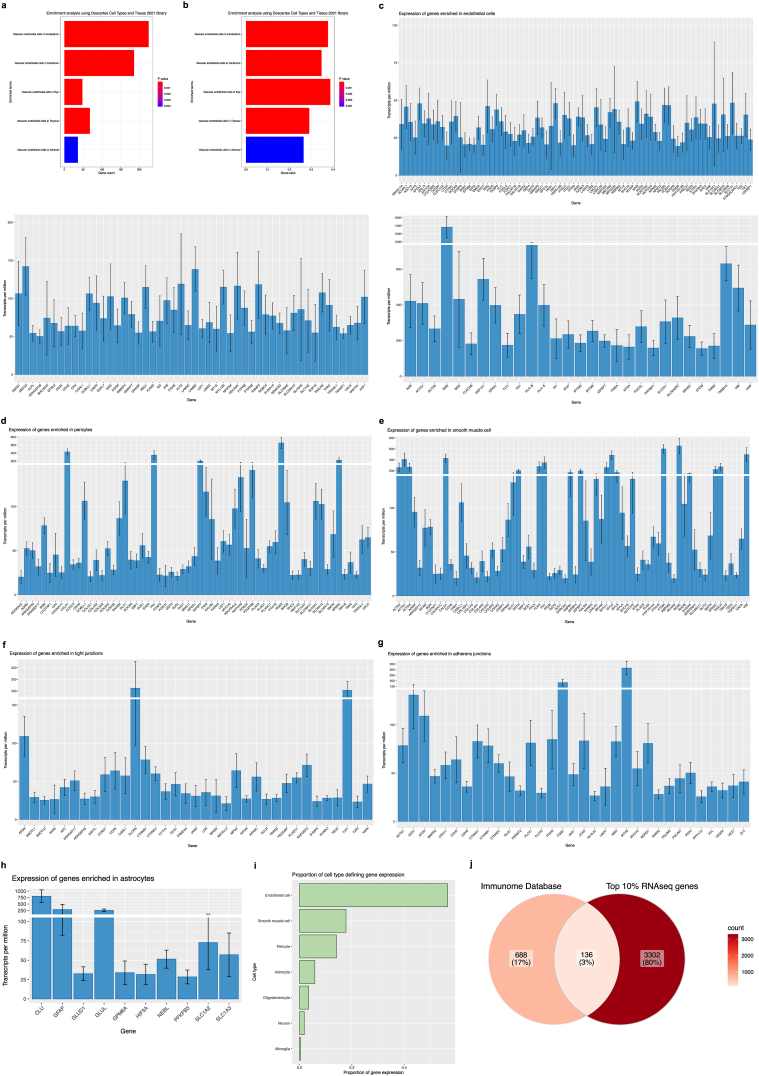
Fig. 6.
a-b) Enrichment analysis of averaged 10% of most highly expressed genes (3437 genes) returns predominantly vascular-related terms. Over-representation analysis (ORA) using the enrichR package in R and the “Descartes Cell Types and Tissue 2021” database was used to identify gene sets that are statistically over-represented. The threshold value of enrichment was selected by a p-value <0.05, indicating that over-represented genes were significantly enriched for vascular-related terms. a) Count for genes in our dataset that are present in returned gene sets. b) Ratio for genes in our dataset that are present in returned gene sets, determined by the total number of genes in each set. c-i) Isolated microvessels have increased expression of canonical neurovascular-related genes. c) Bar plots showing TPMs for endothelial-defining genes, displayed according to general expression range. Highest expression found in endothelial genes B2M, BSG, FLT1, IFITM3, MT2A, SLC2A1, VIM, and VWF. d) Bar plot showing TPMs for pericyte-defining genes. Highest expression was detected in pericyte genes CALD1, FN1, IGFBP7, RGS5, and SPARCL1. e) Bar plot showing TPMs for smooth muscle cell-defining genes. Highest expression was detected in smooth muscle genes ACTG1, ACTN4, MYL6, PTMA, and TAGLN. f) Bar plot showing TPMs for tight junction-defining genes and g) Bar plot showing TPMs for adherens junction-defining genes. Several genes encoding for junctional proteins are found in the top 10% of most highly expressed genes, including CLDN5, CTNNB1, CTNND1, OCLN, JAM1, TJP1, and TJP2. h) Bar plot showing TPMs for astrocyte-defining genes. Astrocytic gene expression was predominantly limited to markers of astrocytic processes or endfeet, namely CLU, GFAP, and GLUL. i) TPMs from neurovascular-related genes were summarized according to cell type expression, demonstrating an overrepresentation of endothelial, smooth muscle cell, and pericyte genes. j) Overlap between top 10% of most highly expressed genes from our RNA sequencing data and immune genes found within the Immunome Database. Bar plots were generated using the ggplot package and Venn diagram was generated using the ggVennDiagram package in R.