Figure 1.
Two mutations in Kmt2d delay early cerebellum development but Kmt2d loss does not drive SHH-MB tumorigenesis
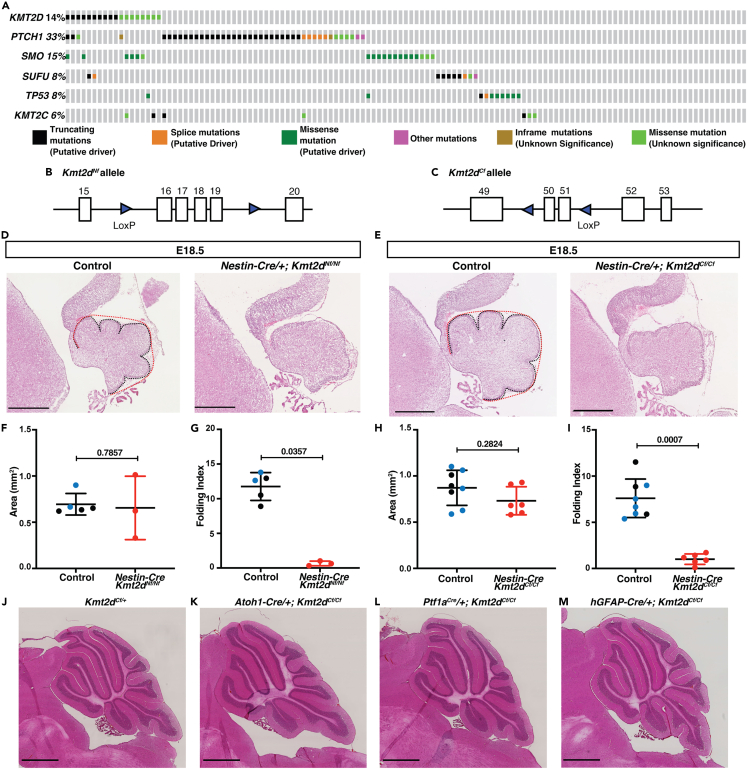
(A) An oncoprint of 127 patient samples considered to be SHH-MB (Kool et al., 2014) using the cBioPortal (Cerami et al., 2012; Gao et al., 2013a) showing the frequency of mutations in KMT2C/D and genes that activate the SHH pathway.
(B) Schematic of mouse Kmt2dNf floxed allele with loxP sites surrounding exons 16–19.63
(C) Schematic of mouse Kmt2dCf floxed allele with loxP sites surrounding exons 50 and 51.64
(D and E) H&E-stained mid-sagittal sections from E18.5 animals of the indicated genotypes with the EGL length in black and convex cerebellar length in red. Scale bars: 500 μm.
(F–I) Quantification at E18.5 of the cerebellar area (mm2) (F and H) and Folding Index ((1-[convex length/EGl length]) x 100) in control (Kmt2dNf/+ and Kmt2dNf/Nf or Kmt2dCf/+ and Kmt2dCf/Cf embryos (black dots) or Nes-Cre/+; Kmt2dNf/+ or Nes-Cre/+; Kmt2dCf/+ (blue dots)) and conditional mutants (red dots) (G and I).
(J–M) Images of H&E-stained mid-sagittal sections from a control and Kmt2dCf/Cf conditional mutants using the three Cre drivers at adult stages (P250-P300). Scale bars: 1 mm. Statistical significance was determined using an unpaired t-test. Error bars: SD. Also see Figure S1.