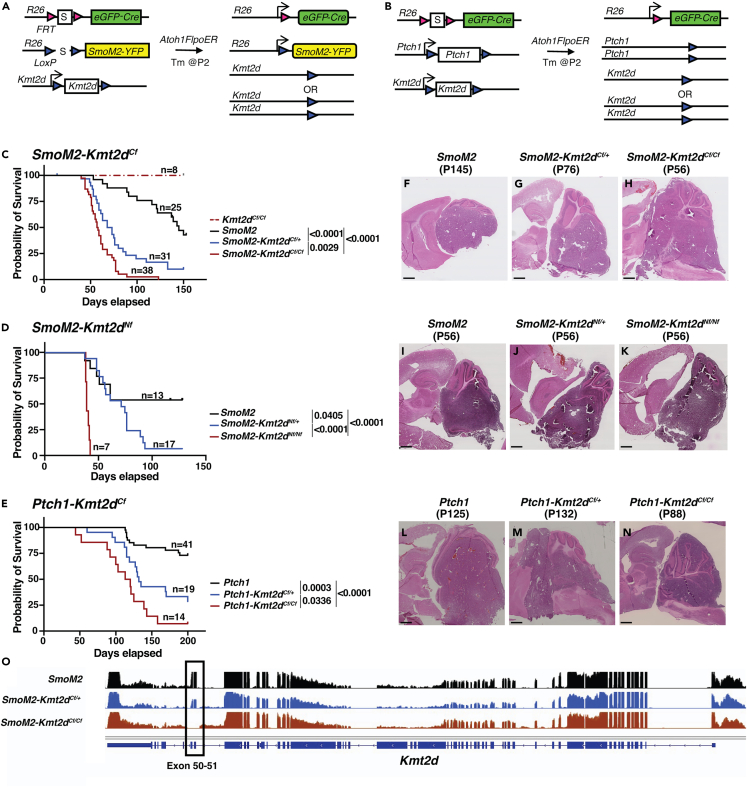
Figure 2.
Two Kmt2d deletion mutations enhance tumor progression in SmoM2-driven SHH-MB and Kmt2d loss also enhances tumor progression when Ptch1 is deleted
(A and B) Schematic showing the two sporadic mouse models of SHH-MB used, SmoM2 (A) and Ptch1fl/fl (B) with and without a heterozygous or homozygous loss of Kmt2d (S: Stop of transcription/translation sequence).
(C–E) Kaplan-Meier curves (left) and statistics (right) for SHH-MB survival using a SmoM2-driven mouse model with a C-terminal (C) or N-terminus (D) heterozygous or homozygous mutation in Kmt2d, and a Ptch1 loss-of-function model with C-terminal mutations of Kmt2d. Statistical significance was calculated using the log-rank test.
(F–N) Representative images of H&E-stained sagittal sections through the hemispheres from end stage SmoM2, SmoM2-Kmt2dCf/+, and SmoM2-Kmt2dCf/Cf animals (F–H), SmoM2, SmoM2-Kmt2dNf/+, and SmoM2-Kmt2dNf/Nf animals (I–K), and Ptch1, Ptch1-Kmt2dCf/+,and Ptch1-Kmt2dCf/Cf animals (L–N). Scale bar: 1 mm.
(O) Overlay of RNA-seq data at the Kmt2d locus in SmoM2 tumors (n = 5), SmoM2-Kmt2dCf/+ tumors (n = 2), and SmoM2-Kmt2dCf/Cf tumors (n = 4). Also see Figure S2.