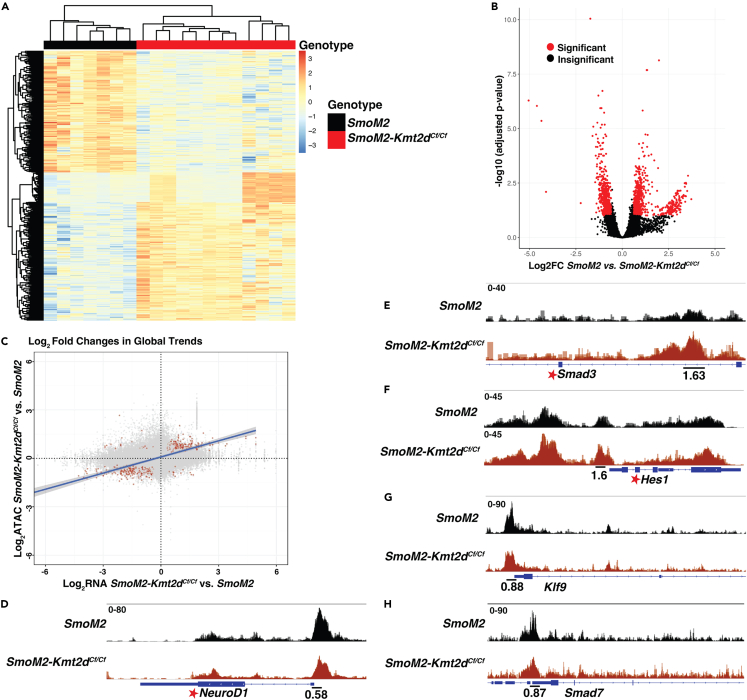
Figure 5.
ATAC-seq data analysis reveals differences in DNA accessibility of pathways associated with neural differentiation and advanced cancer in SmoM2-Kmt2dCf/Cf vs. SmoM2 tumor
(A) Heatmap showing the clustering of differential peaks between SmoM2 and SmoM2-Kmt2dCf/Cf tumors (FC ≥ 1.5, padj ≤ 0.1).
(B) Volcano plots displaying the differentially accessible chromatin regions in SmoM2-Kmt2dCf/Cf compared to SmoM2 tumor cells (FC ≥ 1.5, padj ≤ 0.1).
(C) Comparison of the log2 fold changes (red indicates FC ≥ 1.5, padj ≤ 0.1) in global trends in SmoM2-Kmt2dCf/Cf tumor cells compared to SmoM2 in the RNA-seq and ATAC-seq datasets.
(D–H) ATAC-seq plots showing regions of increased and decreased chromatin accessibility at specific gene loci in SmoM2-Kmt2dCf/Cf (n = 12) compared to SmoM2 (n = 7) primary tumors. The number below indicates the FC in a single 500-bp peak. Fold changes with a padj ≤ 0.1 are indicated with a star. Also see Figure S6 and Tables S5 and S6.