Abstract
Histone variant H3.3 plays novel roles in development as compared to canonical H3 proteins and is the most commonly mutated histone protein of any kind in human disease. Here we discuss how gene targeting studies of the two H3.3-coding genes H3f3a and H3f3b have provided important insights into H3.3 functions including in gametes as well as brain and lung development. Knockouts have also provided insights into the important roles of H3.3 in maintaining genomic stability and chromatin organization, processes that are also affected when H3.3 is mutated in human diseases such as pediatric tumors and neurodevelopmental syndromes. Overall, H3.3 is a unique histone linking development and disease via epigenomic machinery.
Keywords: Histone H3.3, H3f3a, H3f3b, Mouse knockouts
Background
DNA is wrapped around an octamer of histones constituting the nucleosome, which helps to organize and compact the genetic material [1]. A substantial amount of gene regulation occurs at the level of these histones. For example, post-translational modifications (PTMs) of amino acid residues within their N-terminal tails influence the recruitment of transcription factors and other chromatin regulators, as well as the level of compaction of the chromatin at a given gene locus [2–4]. Additionally, multiple histone variants exist and show preferential enrichment at different classes of functional domains, further adding to the complexity of this form of epigenetic regulation [5].
Histone H3.3 is an example of a histone variant with a number of unique properties. Unlike canonical histones, which occur in gene clusters, H3.3 is encoded by just two genes, H3f3a and H3f3b. These two genes also differ from canonical histone genes in that they contain introns and their mRNA is polyadenylated. In contrast to the coordinated gene expression that occurs in histone gene clusters, H3f3a and H3f3b each have unique untranslated regions, promoters, and expression patterns [6–8].
Direct comparisons to canonical H3 proteins reveal that H3.3 differs from H3.1 and H3.2 at 5 and 4 amino acid residues, respectively. Residues that are fully unique to H3.3 are S31, A87, I89, and G90. Phosphorylation of the unique S31 of H3.3 occurs in a number of regulatory contexts and during mitosis [9], while residues 87, 89, and 90 play a role in another important property of H3.3: unlike canonical H3.1 or H3.2, H3.3 can be deposited on chromatin in a replication-independent manner through unique interactions with chaperones HIRA or ATRX and DAXX [10–12]. While replication-independent deposition is an important function of H3.3 in all cells, it takes on a potentially outsized importance in post-mitotic cells, where H3.3 can still be deposited in chromatin, often progressively replacing canonical H3 in nucleosomes over time [13]. Phosphorylation of S31 has several functional roles including at enhancers [14].
Histone variant H3.3 is one of the most highly conserved proteins across eukaryotic species [15]. In fact, the protein sequence is identical in all vertebrates examined. Despite identical amino acid sequences in the H3.3 protein encoded by H3f3a and H3f3b, codon usage differs between the two genes and appears to be under purifying selection [16]. Intriguingly, H3f3a codon usage is more closely aligned with codon usage in proliferation-associated genes, whereas H3f3b codon usage is more comparable to differentiation-associated genes [16], suggesting that over the course of evolution H3f3a and H3f3b may have become optimized for unique transcriptional programs.
H3.3 has a complex role in gene regulation; it is associated with active regions and open chromatin, but is also found at telomeres and repressed genes [11], suggesting context-specific functions in gene regulation and chromosome stability. Despite encoding identical protein sequences, H3f3a and H3f3b appear to have some unique and non-overlapping roles in gene regulation.
Many challenges exist to unraveling the shared and unique roles of histone H3 family members through knockout approaches. These include the fact that multiple variants exist: histone H3 has 5 variants. In addition, in the case of canonical H3 histones like H3.1 and H3.2, there are multiple individual genes arranged in gene clusters to allow for the high level of histone expression needed during S phase [17]. Additionally, many redundancies exist between the different histone H3 variants. Histone proteins are also long-lived, with turnover rates sometimes measured in weeks rather than hours [18–20].
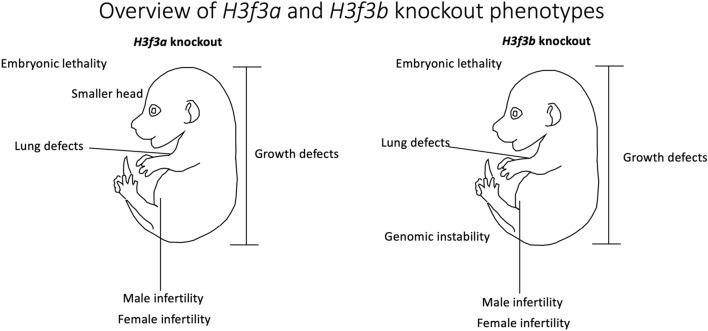
Despite these challenges, several knockout and loss-of-function studies have shed light on the important and distinct roles of H3f3a and H3f3b during development (Table 1). One of the most striking findings was the effect of loss of H3f3a or H3f3b on fertility and gamete formation (Fig. 1). It appears that both genes have vital and non-redundant roles in gamete formation in males, and that H3f3b is also required to varying degrees for gamete formation in females [21–23]. In many other developmental processes, knockout studies indicated that H3f3a and H3f3b were able to compensate for each other, however loss of both resulted in severe growth defects due to genomic instability and activation of the p53 pathway [24]. This role in maintaining the genome appears to be more important than specific gene regulation, as gene expression studies have repeatedly uncovered only modest changes in gene expression with loss of one or both H3.3 genes [22, 25].
Table 1.
Published H3.3 single knockouts and their phenotypes
| Gene knockout | Knockout phenotype | References |
|---|---|---|
| H3f3a, gene trap | 50% neonatal mortality, neuromuscular deficits, reduced fertility in males and females | Couldrey et al. [26] |
| H3f3a, Cre-LoxP | Reduced size, male infertility due to reduced sperm motility and head/tail defects | Tang et al. [23] |
| H3f3a, knockout-first | Almost full embryonic lethality, smaller heads, underdeveloped lungs, some growth defects in heterozygotes | Bush et al. [27] |
| H3f3b, Cre-LoxP (Zp3-Cre) | Partial embryonic lethality, abnormal embryonic development, infertility | Yuen et al. [21] |
| H3f3b, Cre-LoxP (Hprt1-Cre) | Full embryonic/perinatal mortality, growth defects and male infertility in heterozygotes | Tang et al. [23] |
Fig. 1.
Overview of H3f3a and H3f3b knockout phenotypes. Mouse embryo image in diagram was adapted from a Theiler stage reference diagram
Knockout of H3f3a
The first data on knockout of an H3.3 gene in mice came from a gene trap experiment aimed at identifying genes involved in spermatogenesis. Using a LacZ reporter construct, H3f3a was identified as a gene with high expression in the testes [26]. Further characterization revealed high expression in both male and female gonads, specifically in primary spermatocytes and in developing eggs [26].
The gene trap approach resulted in a very strong reduction in H3f3a; levels were below detection by Northern blot, but some H3f3a could still be detected by RT-qPCR. Fifty percent of these homozygous H3f3a mutant pups died shortly after birth, and those that survived showed reduced abilities to compete with their wild-type (WT) littermates, likely due to neuromuscular deficits, as standardized behavior screens showed significant differences between WT and H3f3a mutant mice [26]. Fertility was also found to be reduced in both male and female H3f3a mutant mice, despite reproductive organs appearing normal [26].
Surprisingly, knockout of H3f3a using a Cre-LoxP approach in 129S1/SvImJ (129S1) mice, did not result in the peri-natal lethality observed using the gene trap approach [23]. These H3f3a knockout mice appeared phenotypically normal at birth and survived to adulthood in normal Mendelian ratios, although H3f3a−/− males were smaller than WT littermates at 3 and 6-week timepoints, suggesting a mild potential growth delay [23].
In this study, while H3f3a−/− females had normal fertility, H3f3a−/− males showed signs of reduced fertility and sired smaller litters than WT males [23]. Closer examination revealed lower levels of fertilized ova from the sperm of H3f3a−/− males, likely due to reduced sperm motility as well as head and tail defects [23]. These results indicate that H3f3a is required for spermatogenesis and has a non-redundant role with H3f3b in this process.
A recent paper using a knockout-first approach to disruption of H3f3a observed an even more pronounced phenotype, with almost full embryonic lethality in homozygous null embryos, as well as growth delay in approximately 50% of the heterozygotes [27]. H3f3a knockout embryos had smaller heads and underdeveloped lungs, indicating defects in key developmental processes in the absence of H3f3a.
The finding that knockout of H3f3a resulted in distinct phenotypes in different studies was unexpected: half of pups died perinatally in one study, while another study showed nearly full embryonic lethality, and a third study found no defects in viability. One potential confounding variable that could lead to differences in phenotypes is the longevity of the H3.3 protein itself: even after genetic deletion, H3.3 protein will continue to be present in developing cells for a long period of time, which may influence the severity of the ultimate phenotype. Observed differences in phenotype may also be the result of different background strains of mice used in the various studies or may have to do with the efficiency of the depletion.
The gene-trap approach resulted in very low levels of H3f3a but was not a full knockout [26]. The Cre-LoxP approach to H3f3a knockout used in the second study resulted in excision of the floxed sequence shortly after fertilization of Cre-containing oocytes, and the authors reported very clean deletion with no mosaicism in the resulting offspring [23, 28]. The third study with a knockout first approach that includes the insertion of a large cassette to disrupt the H3f3a locus also resulted in very low but detectable levels of H3f3a mRNA by qPCR and also observed a more severe phenotype [27]. It is a surprising, but never the less a consistent finding, that the two approaches with residual levels of H3f3a mRNA also showed more severe phenotypes than the full Cre deletion. It is possible that insertion of genetic material into the H3f3a locus in the gene trap and knockout-first approaches has a different, and more deleterious effect, than a Cre-based deletion of the locus.
Knockout of H3f3b
The effects of H3f3b loss in mice was studied using a Cre-LoxP system of knockout. H3f3b-floxed mice were crossed with Zp3-Cre to create a germline deletion of H3f3b. Heterozygous (H3f3b+/−) mice were recovered in litters at slightly lower than expected ratios, which may indicate a low level of lethality, while H3f3b−/− mice were recovered at much lower levels than expected, indicating partial lethality [22]. Surviving H3f3b−/− mice were infertile. Examination of embryos at E12.5 showed abnormal development and growth failure in the majority of H3f3b−/− embryos [22].
Furthermore, MEFs isolated from H3f3b−/− mice showed alterations in cell cycle dynamics compared to WT MEFs, and there was a significant increase in chromosomal bridges and abnormal chromosome numbers, suggesting genomic instability in the absence of H3f3b [22]. Staining experiments revealed that knockout MEFs also exhibited increased pericentric heterochromatin and kinetochore proteins, indicating centromere disfunction [22].
Microarray gene expression analysis comparing WT and H3f3b−/− MEFs isolated from littermates at E12.5 showed surprisingly modest changes in overall gene expression as a result of H3f3b knockout [22]. Affected genes were enriched in categories related to histones, DNA synthesis, centromeres, and mitotic regulatory factors. At the protein level, H3S10P, H3K4me3, and H3K9ac levels were reduced [22].
Further studies were completed with these mice to investigate the observed fertility defects of H3f3b loss. Analysis of H3f3b−/− testes revealed abnormal architecture in the seminiferous tubules, and a decrease in sperm concentration as well as sperm abnormalities including decreased motility, high levels of sperm with abnormal heads, and increased apoptosis in the tubules [21], further supporting the idea of an important role for H3.3 proteins in proper chromatin condensation and chromosome segregation. At the epigenetic level, H3K9me3 levels were higher in H3f3b−/− testes compared to WT, while H3K4me3 levels were slightly decreased [21]. H3f3b−/− testes also showed reduced Prm1 staining, indicating that H3f3b plays a role in the switch to protamine-based chromatin during spermatogenesis [21].
Another study employed a different Cre-LoxP approach to knockout H3f3b. H3f3b+/− mice could be generated through a standard Cre-approach, but heterozygous mice were found to be infertile and couldn’t be used to generate knockouts. To get around this, the researchers used a system where Cre is inserted into an X-linked gene (Hprt1), which results in excision of the floxed sequence shortly after fertilization of Cre-containing oocytes in females who are heterozygous for the Cre allele [23]. This study observed more severe phenotypes both in H3f3b+/− and H3f3b−/− compared to the previous study. Using this knockout approach, H3f3b+/− mice were smaller than WT littermates at weaning, and while heterozygous H3f3b+/− females were fertile, males were infertile [23], necessitating the alternative approach to study the effect of homozygous deletion of H3f3b. In approximately half of cases, development of H3f3b−/− embryos did not proceed past the early post-implantation stage [23]. For those embryos that survived, about 24% died between E13 and E18.5, or showed large-scale abnormalities, and the rest died shortly after birth due to respiratory defects [23].
To examine the role of H3f3b in female gametogenesis, a Zp3-Cre line was used to delete H3f3b specifically in the follicle cells [23]. These females had dramatically reduced fertility compared to WT females. This was found to be caused by reduced fertilization of ova, as well as cleavage failure. In these cases, the zygotes did not accumulate high levels of H3Sph, which is a marker of late prophase [23]. When both H3f3a and H3f3b were both conditionally deleted, oocytes were found to be completely inviable [23].
As with H3f3a knockouts, differences in phenotypes obtained using different knockout approaches for H3f3b may be due to the timing of the knockout, the persistence of H3.3 protein coded by H3f3b for a long period of time after knockout, the background strain of the mice, or possibly the mechanism of disruption of the H3f3b locus itself.
H3.3 double knockout
Surprisingly, another study using mice from mixed backgrounds (C57BL/6 and 129) and a Cre-based system to generate germline knockouts of H3f3a and H3f3b found single knockout mice for each gene to be normal and fertile [24]. When the two lines were crossed to generate double knockouts, the embryos usually did not survive past E6.5, suggesting either failure to implant, or to develop further after implantation. In examining combinations of knockout alleles, H3f3a−/− H3f3b+/− mice died perinatally due to breathing problems. H3f3a+/− H3f3b−/− males and females both developed normally [24], however females were fertile while males were infertile due to loss of germ cells.
Sox2-conditional double knockout mice were generated to study the effects of H3.3 loss at a slightly later timepoint during development, as Cre expression starts during the blastocyst stage when under control of the Sox2 promoter. Even with delayed onset of the knockout, Sox2-Cre H3.3 double knockout embryos show delayed growth and an increase in cell death at all timepoints and were reabsorbed by E10.5 [24]. These conditional knockout mice had significantly elevated p21 expression indicating activation of the p53 pathway as a likely cause of the observed cell death and cell cycle arrest phenotype in knockout embryos [24].
Interestingly, and in agreement with previous studies [22, 25], RNA-seq comparing WT and H3.3 knockout embryos revealed only modest effects of loss of H3.3 on gene expression; less than 5% of expressed genes were affected [24]. Among those genes most highly upregulated was the cell cycle inhibitor Cdkn2a [24]. Together, these results suggest that in a p53 WT background, mitotic defects caused by loss of H3.3 activate the p53 pathway, leading to cell cycle arrest and the observed growth defects.
More detailed analysis showed that H3K9me3 and H3K36me2 levels were reduced at telomeres, and heterochromatin domains were characterized by more open chromatin [24]. These differences could disrupt the normal functioning of centromeres and telomeres and thereby contribute to the observed genomic instability in H3.3 knockout mice and cells.
A recent paper studied the effect of H3f3a and H3f3b co-deletion specifically in developing neurons [19]. They knocked out H3f3a and H3f3b in neural progenitor cells using Emx1-Cre, and specifically in excitatory neurons after terminal mitosis using Neuro6d-Cre. In both situations, knockout mice were born alive but died within a few hours of birth [19]. These mice show changes to neuronal fate and identity as well as defects in axon projection and development. Transcriptomic and epigenomic analyses found that accumulation of H3.3 in chromatin is required for properly setting up the transcriptome in newly post-mitotic neurons and regulating the levels of H3K4me3 and H3K27me3 at these genes [19].
H3.3 knockout in ESC
H3.3 double-depletion and double knockout strategies have also been used to study the roles of H3.3 in embryonic stem cells. Jang et al found that H3.3 double knockout ES cells undergo rapid cell death, primarily due to activation of the p53 pathway as a result of mitotic defects [24]. Due to the severe growth defect in these cells that made further characterization difficult, a p53 null mutation was crossed into the conditional knockout lines. With the addition of p53 knockout, the embryos survived until E11.5, and the growth defect was significantly diminished at all timepoints observed [24]. In cell culture, the p53 null background rescues H3.3 knockout cell growth phenotype, however the cells had significantly elevated levels of mitotic defects including lagging chromosomes and anaphase bridges [24]. Comparison of H3.3 knockout MEFs with or without p53 knockout yielded similar results: p53 knockout significantly increased the growth rate of H3.3 knockout MEFs, however these cells had higher levels of mitotic defects and elevated levels of H2A.X S139 phosphorylation, an indication of DNA damage [24].
Another study employed a zinc finger nuclease (ZFN) strategy to knockout both H3f3b alleles, and shRNA against H3f3a in mESC, resulting in strong depletion of total H3.3 in cells. These cells showed reduced nucleosome turnover at active and bivalent genes, and significant depletion of H3K27me3 at bivalent genes [25]. Consistent with mouse studies, RNA-seq analysis revealed only a small group of genes was affected by H3.3 depletion. Notably, they did not observe an effect of loss of the majority of H3.3 on stem cell self-renewal, proliferation, or proper chromosome segregation [25]. Another study with complete H3.3 null ESC found reduced levels of H3K27ac and other acetylation marks at distal enhancers, and determined this to be dependent on phosphorylation of H3.3 at Ser31, a residue unique to histone H3.3 [14].
H3.3 mutations in human disease
Despite producing an identical protein, H3F3A and H3F3B mutations are uniquely associated with several different diseases. In humans, mutations in H3F3A but not H3F3B are strongly associated with pediatric high-grade gliomas, where approximately 70% of tumors bear a lysine to methionine (K27M) mutation in the sequence coding for the N-terminal tail of H3.3, and another 10–15% have a glycine to arginine or valine (G34R/V) mutation in H3F3A [29–31]. These mutations are associated with many gene expression and chromatin changes, as well as genome instability [30, 32]. One of the strongest effects of mutant H3F3A occurs at the level of chromatin structure and epigenetic regulation. Indeed, the H3.3K27M mutation is associated with sharp reductions in the repressive histone modification H3K27me3 and increases in the active modification H3K27ac, as well as the creation of spurious super enhancers at genes with cancer-promoting properties [33–35]. This pattern appears distinct from the situation in H3.3 knockout mice, where loss of H3.3 protein is only correlated with modest changes in histone modifications and gene expression [22].
It is interesting to note that genomic instability is a hallmark of both mutant H3.3 and knockout of H3.3 genes, highlighting important functions for H3.3 in maintaining chromatin structure, regulating chromosome segregation and promoting genome integrity throughout development.
H3.3 mutations are also associated with other human diseases: a K36M mutation specifically in H3F3B is observed in a subset of chondroblastomas, mutations of H3F3A are found in giant cell tumors of the bone, and overexpression of H3F3A in lung cancer leads to aberrant histone deposition and activation of metastasis-associated genes [36, 37].
At the same time, H3f3a and H3f3b do appear to share many functions. In one example, functionally similar or identical de novo germline mutations in either H3F3A or H3F3B have recently been identified in patients with a rare neurodegenerative disorder [38]. Patients with this condition suffer from developmental delay, epilepsy, neurodegeneration, and, in some cases, congenital abnormalities [38]. The mutations found in this condition are predicted to disrupt core H3.3 functions, including interactions with DNA, with other histones in the nucleosome, or with chaperone proteins, which may explain why mutations in H3F3A or H3F3B can lead to similar or even the same phenotypes [38]. When either gene is uniquely associated with a particular disease, it is possible that is explained by their unique expression patterns in distinct cells or the chromatin at either locus conferring more susceptibility to mutations.
H3.3 depletion in other species
H3.3 loss of function has also been studied in other species. In Caenorhabditis elegans H3.3 is highly expressed and incorporated into chromatin throughout development and during adulthood, with the highest levels of H3.3 observed in post-mitotic cells. H3.3-deficient animals developed normally and are fertile, however they had lower survival rates when exposed to stress [39]. H3.3 loss in Xenopus laevis caused developmental defects during late gastrulation including spina bifida, open blastopore, and shortened anteroposterior axis [40].
In Drosophila, where H3.3 makes up approximately 25% of the total H3 protein in the cell, single H3.3 knockouts where phenotypically normal, but double knockouts were infertile and had reduced viability [41]. Examination of the testes of H3.3-double-deficient flies showed defects in chromosome condensation and segregation during meiosis, with lagging chromosomes frequently observed in Anaphase I and chromosome bridges in Anaphase II, suggesting a crucial role for H3.3 in chromosome organization and condensation during gamete production [41]. These defects can be rescued by ectopic expression of the canonical histone H3.2 [41], suggesting the total level of histone H3 could be more important than the specific variant being expressed in some contexts. The infertility and chromosome segregation defects observed in H3.3 deficient Drosophila are similar to defects seen in H3.3 knockout mice and suggest an evolutionarily conserved role for H3.3 in fertility and, more specifically, chromosome segregation during gamete production.
H3.3 chaperone knockouts
Specific chaperone proteins are required for the incorporation of histones into chromatin. For example, the HIRA chaperone complex is responsible for depositing H3.3 into chromatin at active regions, while the ATRX/DAXX complex incorporates H3.3 at telomeres and pericentromeric heterochromatin. Hira knockout results in embryonic lethality around E10.5, with multiple major developmental abnormalities indicative of patterning defects originating during gastrulation [42].
Similar to H3.3, HIRA plays an important role in gamete generation. When Hira is specifically knocked-out during oogenesis using a Zp3-Cre system, oocytes develop normally, and females undergo ovulation, but are infertile [43]. Further examination indicated that Hira knockout in the oocyte causes zygotes to fail to reach the 2-cell stage, even if the sperm are from WT males [43]. Fitting with the observation that after fertilization the male pronucleus appears devoid of histones in Hira mutants, is the idea that H3.3 and its chaperone together have a unique role in histone deposition at this timepoint. Sperm DNA mostly has protamines rather than conventional histones, and needs to be repackaged after fertilization occurs [43]. Since this repackaging occurs before the first cell cycle, only a cell cycle-independent H3 variant in the form of H3.3 can be used for this process, and HIRA is a crucial part of the H3.3 deposition pathway.
Loss of function of the ATRX/DAXX chaperone complex, which is responsible for H3.3 deposition at heterochromatic and pericentromeric regions, results in a number of defects. Atrx knockout mESC cell lines have increased levels of DNA damage compared to WT mESC, and delayed progression through S phase, due to replication fork stalling at repetitive regions and sites of heterochromatin, where ATRX is known to bind [44]. Similar S phase delay, DNA damage, and telomere fragility are also observed in Atrx conditional knockout in myoblasts [45] and neural progenitor cells [46]. Daxx knockout also results in early embryonic lethality, around day E9.5. Daxx−/− embryos are smaller than WT littermates and also appear highly disorganized [47]. Cells isolated from these mice show increased rates of apoptosis compared to WT controls, possibly due to defects in DNA damage response or other related pathways [47].
Knockout of histone variants with functions related to H3.3
There is some general overlap in knockout phenotypes for variants H2AZ and H2AX with H3.3 and ATRX knockouts. For example, H2AX knockout mice are born at expected ratios, but have a growth defect compared to their WT littermates [48]. This phenotype is at least partially attributable to cell cycle arrest, due to the key role of H2AX phosphorylation in the DNA damage response [48]. Like H3.3, H2AX plays an important role in genome stability, highlighted by the finding that H2AX−/− MEFs have a significant increase in karyotypic abnormalities including chromatid breaks and dicentric chromosomes. H2AX−/− female mice are sub-fertile, while H2AX−/− male mice are completely infertile, with thinner seminiferous tubules and no detectable mature sperm [48].
Histone H2AZ is another variant of histone H2 that shares some similarities with H3.3. It is also encoded by 2 genes (H2az.1 and H2az.2) although lacking introns. H2AZ is expressed throughout the cell cycle, and is enriched at euchromatic regions as well as at heterochromatin and centromeres. Indeed, H2AZ and H3.3 are often found together in “double variant” nucleosomes that have higher levels of instability, contributing to high nucleosome turnover and open chromatin in these regions [49]. H2AZ knockout mice die early in development [50], and siRNA knockdown of H2AZ results in major chromosomal abnormalities including lagging chromosomes and the presence of chromatin bridges between dividing nuclei [51]. These effects are at least in part due to the disruption of HP1α binding at heterochromatin [51]. The overlap in fertility, growth, and chromosome segregation phenotypes between different histone variant knockouts underscores the importance of histone variants in proper chromosome segregation during processes like gametogenesis, as well as the many rounds of mitosis required for proper growth and development of an organism.
Conclusions
While knockout and depletion studies on histones can be challenging due to the many histone genes and proteins, the presence of variants, and the long half-lives of these proteins, a number of recent papers have successfully employed these strategies to better understand the roles of H3.3. These have identified key roles for histone H3.3 in genome integrity, including proper segregation of chromosomes during cell division, and crucial and non-redundant roles of H3f3a and H3f3b in gamete formation in mice, highlighting the important and unique regulatory roles of histone variants like H3.3 (Fig. 1). The relatively mild effects of loss of H3.3 on the transcriptome have been consistently observed, suggesting either a lesser role in gene expression or complex redundancies or compensatory mechanisms. Overall, H3.3 knockout studies provide important context for understanding the mechanisms of H3.3 mutations in cancers and brain development, and may uncover roles for H3.3 in additional tissues and diseases in the future.
Acknowledgements
This work was supported by the following NIH grants to PSK: 1R01GM116919 and 1R01NS106878.
Abbreviations
- WT
Wild type
- MEF
Mouse embryonic fibroblast
Author contributions
RHK and PK wrote and edited the manuscript.
Funding
Not applicable.
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Stillman B. Histone modifications: insights into their influence on gene expression. Cell. 2018;175(1):6–9. doi: 10.1016/j.cell.2018.08.032. [DOI] [PubMed] [Google Scholar]
- 3.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang T, Cooper S, Brockdorff N. The interplay of histone modifications—writers that read. EMBO Rep. 2015;16(11):1467–1481. doi: 10.15252/embr.201540945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Talbert PB, Henikoff S. Histone variants at a glance. J Cell Sci. 2021;134(6):jcs244749. doi: 10.1242/jcs.244749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frank D, Doenecke D, Albig W. Differential expression of human replacement and cell cycle dependent H3 histone genes. Gene. 2003;312:135–143. doi: 10.1016/S0378-1119(03)00609-7. [DOI] [PubMed] [Google Scholar]
- 7.Akhmanova AS, Bindels PC, Xu J, Miedema K, Kremer H, Hennig W. Structure and expression of histone H3.3 genes in Drosophila melanogaster and Drosophila hydei. Genome. 1995;38(3):586–600. doi: 10.1139/g95-075. [DOI] [PubMed] [Google Scholar]
- 8.Krimer DB, Cheng G, Skoultchi AI. Induction of H33 replacement histone mRNAs during the precommitment period of murine erythroleukemia cell differentiation. Nucleic Acids Res. 1993;21(12):2873–2879. doi: 10.1093/nar/21.12.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hake SB, Garcia BA, Kauer M, Baker SP, Shabanowitz J, Hunt DF, et al. Serine 31 phosphorylation of histone variant H3.3 is specific to regions bordering centromeres in metaphase chromosomes. Proc Natl Acad Sci U S A. 2005;102(18):6344–6349. doi: 10.1073/pnas.0502413102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9(6):1191–1200. doi: 10.1016/S1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 11.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140(5):678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci U S A. 2010;107(32):14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tvardovskiy A, Schwämmle V, Kempf SJ, Rogowska-Wrzesinska A, Jensen ON. Accumulation of histone variant H3.3 with age is associated with profound changes in the histone methylation landscape. Nucleic Acids Res. 2017;45(16):9272–9289. doi: 10.1093/nar/gkx696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martire S, Gogate AA, Whitmill A, Tafessu A, Nguyen J, Teng YC, et al. Phosphorylation of histone H3.3 at serine 31 promotes p300 activity and enhancer acetylation. Nat Genet. 2019;51(6):941–946. doi: 10.1038/s41588-019-0428-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malik HS, Henikoff S. Phylogenomics of the nucleosome. Nat Struct Biol. 2003;10(11):882–891. doi: 10.1038/nsb996. [DOI] [PubMed] [Google Scholar]
- 16.Muhire BM, Booker MA, Tolstorukov MY. Non-neutral evolution of H3.3-encoding genes occurs without alterations in protein sequence. Sci Rep. 2019;9(1):8472. doi: 10.1038/s41598-019-44800-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szenker E, Ray-Gallet D, Almouzni G. The double face of the histone variant H3.3. Cell Res. 2011;21(3):421–434. doi: 10.1038/cr.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zee BM, Levin RS, DiMaggio PA, Garcia BA. Global turnover of histone post-translational modifications and variants in human cells. Epigenet Chromatin. 2010;3(1):22. doi: 10.1186/1756-8935-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Funk OH, Qalieh Y, Doyle DZ, Lam MM, Kwan KY. Postmitotic accumulation of histone variant H3.3 in new cortical neurons establishes neuronal chromatin, transcriptome, and identity. Proc Natl Acad Sci U S A. 2022;119(32):e2116956119. doi: 10.1073/pnas.2116956119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toyama BH, Savas JN, Park SK, Harris MS, Ingolia NT, Yates JR, 3rd, et al. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell. 2013;154(5):971–982. doi: 10.1016/j.cell.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuen BT, Bush KM, Barrilleaux BL, Cotterman R, Knoepfler PS. Histone H3.3 regulates dynamic chromatin states during spermatogenesis. Development. 2014;141(18):3483–3494. doi: 10.1242/dev.106450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bush KM, Yuen BT, Barrilleaux BL, Riggs JW, O'Geen H, Cotterman RF, et al. Endogenous mammalian histone H3.3 exhibits chromatin-related functions during development. Epigenet Chromatin. 2013;6(1):7. doi: 10.1186/1756-8935-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang MC, Jacobs SA, Mattiske DM, Soh YM, Graham AN, Tran A, et al. Contribution of the two genes encoding histone variant h3.3 to viability and fertility in mice. PLoS Genet. 2015;11(2):e1004964. doi: 10.1371/journal.pgen.1004964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jang CW, Shibata Y, Starmer J, Yee D, Magnuson T. Histone H3.3 maintains genome integrity during mammalian development. Genes Dev. 2015;29(13):1377–1392. doi: 10.1101/gad.264150.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banaszynski LA, Wen D, Dewell S, Whitcomb SJ, Lin M, Diaz N, et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell. 2013;155(1):107–120. doi: 10.1016/j.cell.2013.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Couldrey C, Carlton MB, Nolan PM, Colledge WH, Evans MJ. A retroviral gene trap insertion into the histone 3.3A gene causes partial neonatal lethality, stunted growth, neuromuscular deficits and male sub-fertility in transgenic mice. Hum Mol Genet. 1999;8(13):2489–2495. doi: 10.1093/hmg/8.13.2489. [DOI] [PubMed] [Google Scholar]
- 27.Bush K, Cervantes V, Yee JQ, Klein RH, Knoepfler PS. A knockout-first model of H3f3a gene targeting leads to developmental lethality. Genesis. 2023;61(1–2):e23507. doi: 10.1002/dvg.23507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang MC, Jacobs SA, Wong LH, Mann JR. Conditional allelic replacement applied to genes encoding the histone variant H3.3 in the mouse. Genesis. 2013;51(2):142–146. doi: 10.1002/dvg.22366. [DOI] [PubMed] [Google Scholar]
- 29.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 30.Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44(3):251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuen BT, Knoepfler PS. Histone H3.3 mutations: a variant path to cancer. Cancer Cell. 2013;24(5):567–574. doi: 10.1016/j.ccr.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bočkaj I, Martini TEI, de Camargo Magalhães ES, Bakker PL, Meeuwsen Boer TGJ, Armandari I, et al. The H3.3K27M oncohistone affects replication stress outcome and provokes genomic instability in pediatric glioma. PLoS Genet. 2021;17(11):e1009868. doi: 10.1371/journal.pgen.1009868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen KY, Bush K, Klein RH, Cervantes V, Lewis N, Naqvi A, et al. Reciprocal H3.3 gene editing identifies K27M and G34R mechanisms in pediatric glioma including NOTCH signaling. Commun Biol. 2020;3(1):363. doi: 10.1038/s42003-020-1076-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagaraja S, Vitanza NA, Woo PJ, Taylor KR, Liu F, Zhang L, et al. Transcriptional dependencies in diffuse intrinsic pontine glioma. Cancer Cell. 2017;31(5):635–52.e6. doi: 10.1016/j.ccell.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis PW, Müller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340(6134):857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park SM, Choi EY, Bae M, Kim S, Park JB, Yoo H, et al. Histone variant H3F3A promotes lung cancer cell migration through intronic regulation. Nat Commun. 2016;7:12914. doi: 10.1038/ncomms12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Behjati S, Tarpey PS, Presneau N, Scheipl S, Pillay N, Van Loo P, et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet. 2013;45(12):1479–1482. doi: 10.1038/ng.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bryant L, Li D, Cox SG, Marchione D, Joiner EF, Wilson K, et al. Histone H3.3 beyond cancer: Germline mutations in. Sci Adv. 2020;6(49):eabc9207. doi: 10.1126/sciadv.abc9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strobino M, Wenda JM, Padayachy L, Steiner FA. Loss of histone H3.3 results in DNA replication defects and altered origin dynamics in. Genome Res. 2020;30(12):1740–1751. doi: 10.1101/gr.260794.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szenker E, Lacoste N, Almouzni G. A developmental requirement for HIRA-dependent H3.3 deposition revealed at gastrulation in Xenopus. Cell Rep. 2012;1(6):730–740. doi: 10.1016/j.celrep.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 41.Sakai A, Schwartz BE, Goldstein S, Ahmad K. Transcriptional and developmental functions of the H3.3 histone variant in Drosophila. Curr Biol. 2009;19(21):1816–1820. doi: 10.1016/j.cub.2009.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts C, Sutherland HF, Farmer H, Kimber W, Halford S, Carey A, et al. Targeted mutagenesis of the Hira gene results in gastrulation defects and patterning abnormalities of mesoendodermal derivatives prior to early embryonic lethality. Mol Cell Biol. 2002;22(7):2318–2328. doi: 10.1128/MCB.22.7.2318-2328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin CJ, Koh FM, Wong P, Conti M, Ramalho-Santos M. Hira-mediated H3.3 incorporation is required for DNA replication and ribosomal RNA transcription in the mouse zygote. Dev Cell. 2014;30(3):268–279. doi: 10.1016/j.devcel.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clynes D, Jelinska C, Xella B, Ayyub H, Taylor S, Mitson M, et al. ATRX dysfunction induces replication defects in primary mouse cells. PLoS ONE. 2014;9(3):e92915. doi: 10.1371/journal.pone.0092915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huh MS, Price O'Dea T, Ouazia D, McKay BC, Parise G, Parks RJ, et al. Compromised genomic integrity impedes muscle growth after Atrx inactivation. J Clin Invest. 2012;122(12):4412–4423. doi: 10.1172/JCI63765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bérubé NG, Mangelsdorf M, Jagla M, Vanderluit J, Garrick D, Gibbons RJ, et al. The chromatin-remodeling protein ATRX is critical for neuronal survival during corticogenesis. J Clin Invest. 2005;115(2):258–267. doi: 10.1172/JCI200522329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Michaelson JS, Bader D, Kuo F, Kozak C, Leder P. Loss of Daxx, a promiscuously interacting protein, results in extensive apoptosis in early mouse development. Genes Dev. 1999;13(15):1918–1923. doi: 10.1101/gad.13.15.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296(5569):922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21(12):1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Faast R, Thonglairoam V, Schulz TC, Beall J, Wells JR, Taylor H, et al. Histone variant H2A.Z is required for early mammalian development. Curr Biol. 2001;11(15):1183–1187. doi: 10.1016/S0960-9822(01)00329-3. [DOI] [PubMed] [Google Scholar]
- 51.Rangasamy D, Greaves I, Tremethick DJ. RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nat Struct Mol Biol. 2004;11(7):650–655. doi: 10.1038/nsmb786. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.