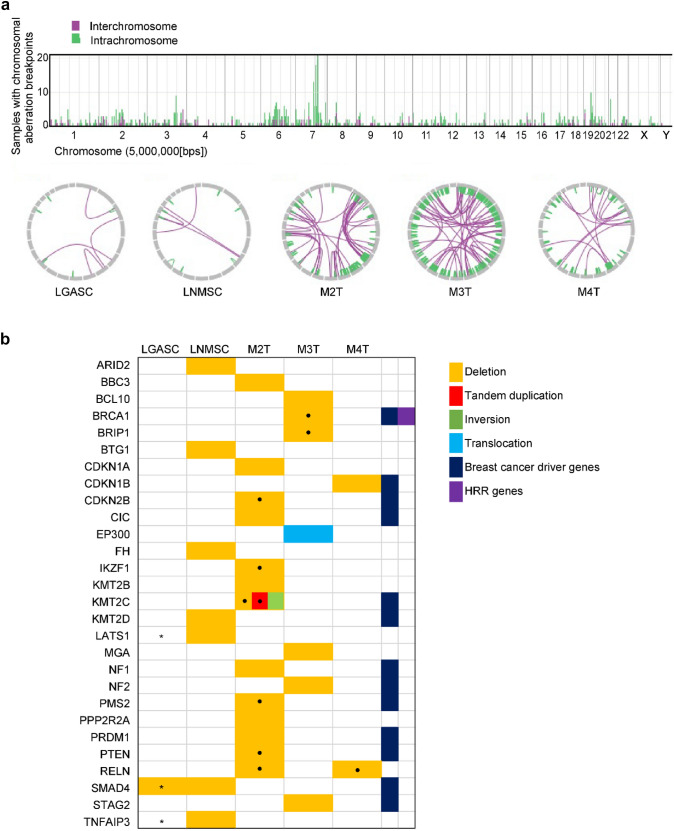
Fig. 3.
Summary of the SVs of the five metaplastic carcinomas. a The bar graph in the upper part represents the number of breakpoints of SVs per chromosome for the five samples. The Circos plots in the lower part show two breakpoints connected by a line for each metaplastic carcinoma. b The genes affected by SVs (deletion or tandem duplication or fusion) were curated as oncogenic or likely oncogenic in the OncoKB database. The type of SV is color-coded as stated in the legend. The dot in the box indicates that multiple SVs are identified in the corresponding genes. Asterisks in the boxes of LGASC indicate that LGASC has few reads representing the same deletion as LNMSC confirmed using Integrative Genomics Viewer, which is not recognized by the Genomon 2 DNA analysis pipeline because of the threshold. Namely, LGASC not only showed SMAD4 deletion in intron 1 but also a read indicating a SMAD4::DCC fusion gene identical to that found in LNMSC. SV, structural variation; LGASC, low-grade adenosquamous carcinoma; LNMSC, lymph node metastasis consisting of high-grade metaplastic carcinoma of the breast with a predominant metaplastic squamous cell carcinoma component; HRR, homologous recombinational repair