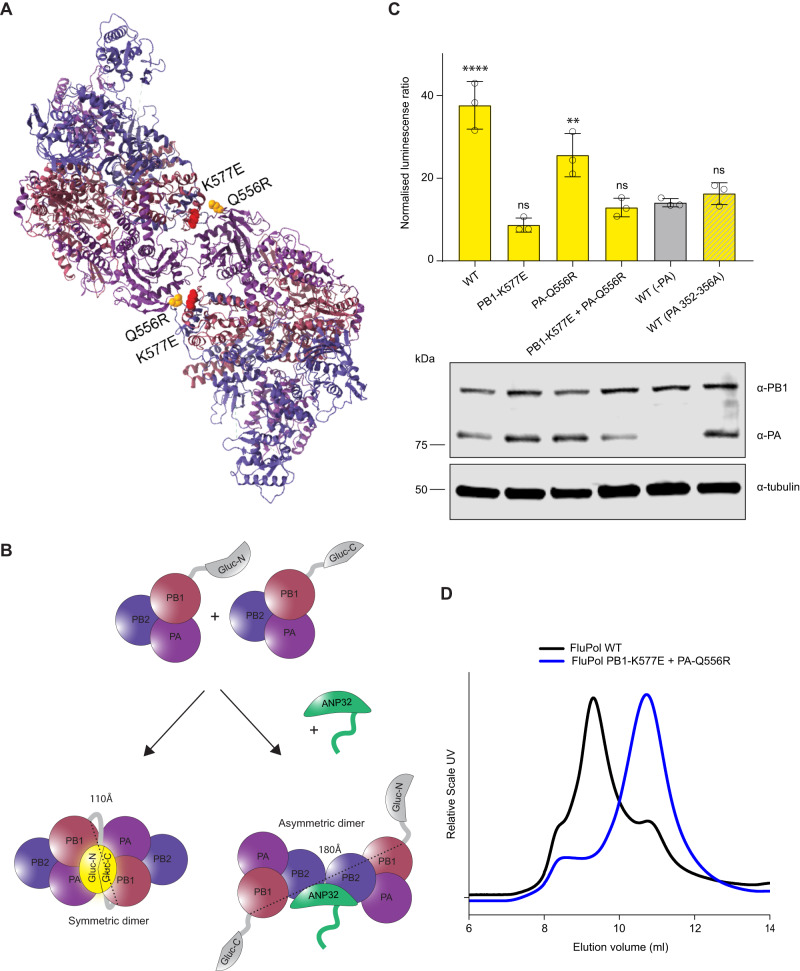
Fig. 5. PB1 K577E destabilises formation of the polymerase dimer.
A Symmetric dimer of influenza polymerase (PDB: 6QX8) showing PB1 K577E in red and PA Q556R in yellow. B Schematic showing the split-luciferase dimerization assay with C-terminus of Gaussia luciferase and the N-terminus of Gaussia luciferase attached to separate PB1 proteins. The two halves of the luciferase are 110Å in the same plane of the symmetric dimer and 180 Å on opposite sides of the asymmetric dimer. C Split luciferase complementation assay measuring dimerization of Tky05 WT, PB1 K577E + PA Q556R, PB1 K577E and PA Q556R polymerases (formed using equal amounts of PB1-Gluc1 + PB1-Gluc2 or PB1-K577E-Gluc1 + PB1-K577E-Gluc2). Data presented are representative of n = 3 biological repeats each conducted with n = 3 wells, presented as mean values +/− SD. Statistical significance was determined by multiple comparisons of one-way ANOVA, samples were compared to WT (-PA), *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (-PA vs WT p < 0.0001, -PA vs PB1-K577E p = 0.2708, -PA vs PA-Q556R p = 0.0019, -PA vs PB1-K577E + PA-Q556R p = 0.9993, -PA vs WT(PA 352-356A) p = 0.9683). Accompanying Western blot showing expression of PB1/K577E-Gluc, PA/Q556R and tubulin. D Tky05 WT polymerase and PB1 K577E + PA Q556R were cloned into baculovirus and expressed in Sf9 cells. The polymerases were purified and concentrated (as described in the methods) then analysed by size exclusion chromatography. Lower elution volume is indicative of higher molecular weight. Source data are provided as a Source Data file.