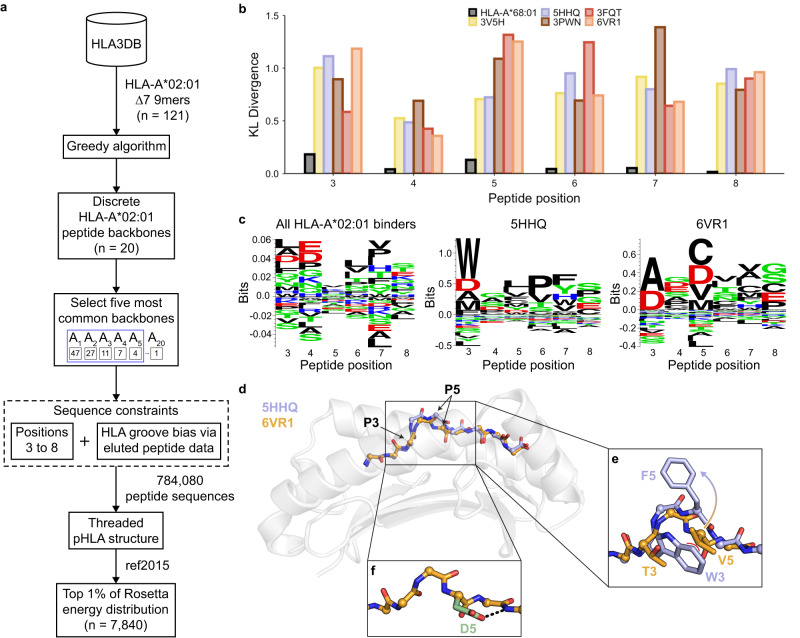
Fig. 3. Exhaustive enumeration of peptide sequence space reveals biases imposed by different backbone conformations.
a A schematic of the exhaustive structural modeling conducted to augment the existing peptide sequence space for HLA-A*02:01. b Kullback–Leibler (KL) divergence between each of the predicted sequence space of the five most common HLA-A*02:01 backbones represented by PDB IDs and HLA-A*02:01 reference sequence space. A comparison between the eluted peptides bound to HLA-A*68:01 and those bound to HLA-A*02:01 was included as a negative control. c Peptide sequence logos of all eluted peptides shown to bind to HLA-A*02:01 via the IEDB (n = 32,483) and from structural modeling results (n = 7840). Created using Seq2Logo90. d Structural overlay of the peptide backbones (5HHQ [10.2210/pdb5HHQ/pdb], blue and 6VR1 [10.2210/pdb6VR1/pdb], orange) bound to HLA-A*02:01 (gray) with the peptide Cα atoms shown as spheres. e An enlarged view of the middle of the peptide with side chains depicted for P3 and P5. Coloring of the peptide backbone and side chain is identical to (d). Steric hindrance is shown as a red curve. f Structural model of the 6VR1 [10.2210/pdb6VR1/pdb] peptide backbone with P5 mutated to aspartate, as shown in green. A hydrogen bond is shown as a dashed black line.