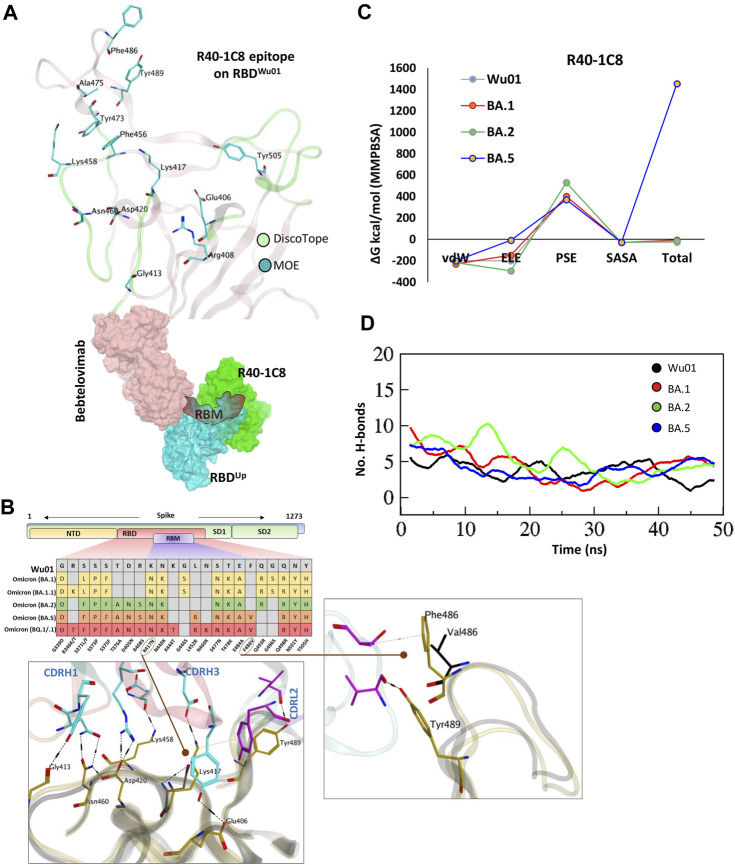
FIGURE 3.
The epitope mapping and escape of BA.5, BQ.1.1, and XBB.1.5 subvariants from R40-1C8. (A) The epitope residues on RBD are predicted through DiscoTope (green color cartoon representation) and Molecular Operating Environment (MOE, Cyan color). The surface maps (colored according to chains) shows that R40-1C8 does not compete with Bebtelovimab. (B) Mutations in the RBD region of Spike protein of SARS-CoV-2 Omicron variants and their effect on the interface contacts with respect to R40-1C8. Mutations [including Phe(F)486Val(V)] at the interface are indicated with stick and circles. (C) Changes in the binding free energy of R40-1C8 bound to the SARS-CoV-2 variants. (D) Changes in the number of Hydrogen bonds between R40-1G8 and respective variants.