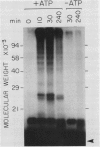
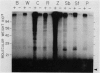
Abstract
Ubiquitin is a highly conserved, 76-amino acid polypeptide with several important regulatory functions in both plants and animals that all arise from its covalent ligation to other cellular proteins. Here, we demonstrate that higher plants have the capacity to conjugate ubiquitin to other plant proteins in vitro. Using 125I-labeled human ubiquitin as a substrate, conjugating activities were observed in crude etiolated tissue extracts from all species tested, including oats, rye, barley, corn, zucchini squash, pea, soybean, and sunflower. The reaction has a soluble distribution, is specific for ATP, and requires the protease inhibitor, leupeptin, to protect ubiquitin from inactivation during the assay. Conjugation is inhibited by N-ethylmaleimide and high concentrations of 2-mercaptoethanol suggesting that the mechanism of ubiquitin ligation in plants involves a similar thiolester intermediate to that found in the mammalian pathway. The conjugating activity in etiolated oat extracts is extremely labile with a half-life of about 20 minutes at 30°C. Detectable but low ATP-stimulated, conjugating activities were also observed in extracts from dry seeds and green leaves of oats. In addition to this conjugating activity, crude plant extracts have the capacity to degrade ubiquitin-protein conjugates formed in vitro. These results demonstrate that higher plants contain several of the enzymic activities necessary for ubiquitin's functions and provide a method for assaying ubiquitin conjugation in vitro.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bensadoun A., Weinstein D. Assay of proteins in the presence of interfering materials. Anal Biochem. 1976 Jan;70(1):241–250. doi: 10.1016/s0003-2697(76)80064-4. [DOI] [PubMed] [Google Scholar]
- Boller T., Kende H. Hydrolytic enzymes in the central vacuole of plant cells. Plant Physiol. 1979 Jun;63(6):1123–1132. doi: 10.1104/pp.63.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canut H., Alibert G., Boudet A. M. Hydrolysis of Intracellular Proteins in Vacuoles Isolated from Acer pseudoplatanus L. Cells. Plant Physiol. 1985 Dec;79(4):1090–1093. doi: 10.1104/pp.79.4.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A., Finley D., Varshavsky A. Ubiquitin dependence of selective protein degradation demonstrated in the mammalian cell cycle mutant ts85. Cell. 1984 May;37(1):57–66. doi: 10.1016/0092-8674(84)90300-3. [DOI] [PubMed] [Google Scholar]
- Ciechanover A., Heller H., Elias S., Haas A. L., Hershko A. ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1365–1368. doi: 10.1073/pnas.77.3.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A., Heller H., Katz-Etzion R., Hershko A. Activation of the heat-stable polypeptide of the ATP-dependent proteolytic system. Proc Natl Acad Sci U S A. 1981 Feb;78(2):761–765. doi: 10.1073/pnas.78.2.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox M. J., Shapira R., Wilkinson K. D. Tryptic peptide mapping of ubiquitin and derivatives using reverse-phase high performance liquid chromatography. Anal Biochem. 1986 Apr;154(1):345–352. doi: 10.1016/0003-2697(86)90535-x. [DOI] [PubMed] [Google Scholar]
- Etlinger J. D., Goldberg A. L. A soluble ATP-dependent proteolytic system responsible for the degradation of abnormal proteins in reticulocytes. Proc Natl Acad Sci U S A. 1977 Jan;74(1):54–58. doi: 10.1073/pnas.74.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D., Ciechanover A., Varshavsky A. Thermolability of ubiquitin-activating enzyme from the mammalian cell cycle mutant ts85. Cell. 1984 May;37(1):43–55. doi: 10.1016/0092-8674(84)90299-x. [DOI] [PubMed] [Google Scholar]
- Goldberg A. L., St John A. C. Intracellular protein degradation in mammalian and bacterial cells: Part 2. Annu Rev Biochem. 1976;45:747–803. doi: 10.1146/annurev.bi.45.070176.003531. [DOI] [PubMed] [Google Scholar]
- Goldknopf I. L., Busch H. Isopeptide linkage between nonhistone and histone 2A polypeptides of chromosomal conjugate-protein A24. Proc Natl Acad Sci U S A. 1977 Mar;74(3):864–868. doi: 10.1073/pnas.74.3.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldknopf I. L., Wilson G., Ballal N. R., Busch H. Chromatin conjugate protein A24 is cleaved and ubiquitin is lost during chicken erythropoiesis. J Biol Chem. 1980 Nov 25;255(22):10555–10558. [PubMed] [Google Scholar]
- Goldstein G., Scheid M., Hammerling U., Schlesinger D. H., Niall H. D., Boyse E. A. Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc Natl Acad Sci U S A. 1975 Jan;72(1):11–15. doi: 10.1073/pnas.72.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas A. L., Murphy K. E., Bright P. M. The inactivation of ubiquitin accounts for the inability to demonstrate ATP, ubiquitin-dependent proteolysis in liver extracts. J Biol Chem. 1985 Apr 25;260(8):4694–4703. [PubMed] [Google Scholar]
- Haas A. L., Wilkinson K. D. The large scale purification of ubiquitin from human erythrocytes. Prep Biochem. 1985;15(1-2):49–60. doi: 10.1080/00327488508062433. [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A., Heller H., Haas A. L., Rose I. A. Proposed role of ATP in protein breakdown: conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1783–1786. doi: 10.1073/pnas.77.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. Mechanisms of intracellular protein breakdown. Annu Rev Biochem. 1982;51:335–364. doi: 10.1146/annurev.bi.51.070182.002003. [DOI] [PubMed] [Google Scholar]
- Hershko A., Eytan E., Ciechanover A., Haas A. L. Immunochemical analysis of the turnover of ubiquitin-protein conjugates in intact cells. Relationship to the breakdown of abnormal proteins. J Biol Chem. 1982 Dec 10;257(23):13964–13970. [PubMed] [Google Scholar]
- Hershko A., Eytan E., Ciechanover A., Haas A. L. Immunochemical analysis of the turnover of ubiquitin-protein conjugates in intact cells. Relationship to the breakdown of abnormal proteins. J Biol Chem. 1982 Dec 10;257(23):13964–13970. [PubMed] [Google Scholar]
- Hershko A., Heller H., Elias S., Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983 Jul 10;258(13):8206–8214. [PubMed] [Google Scholar]
- Hershko A., Leshinsky E., Ganoth D., Heller H. ATP-dependent degradation of ubiquitin-protein conjugates. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1619–1623. doi: 10.1073/pnas.81.6.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzer H., Heinrich P. C. Control of proteolysis. Annu Rev Biochem. 1980;49:63–91. doi: 10.1146/annurev.bi.49.070180.000431. [DOI] [PubMed] [Google Scholar]
- Hough R., Pratt G., Rechsteiner M. Ubiquitin-lysozyme conjugates. Identification and characterization of an ATP-dependent protease from rabbit reticulocyte lysates. J Biol Chem. 1986 Feb 15;261(5):2400–2408. [PubMed] [Google Scholar]
- Hough R., Rechsteiner M. Ubiquitin-lysozyme conjugates. Purification and susceptibility to proteolysis. J Biol Chem. 1986 Feb 15;261(5):2391–2399. [PubMed] [Google Scholar]
- Kahl G., Stegemann H. Enzyme degradation in higher plants: phosphoglucomutase. FEBS Lett. 1973 Jun 1;32(2):325–329. doi: 10.1016/0014-5793(73)80865-8. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Levinger L., Varshavsky A. Selective arrangement of ubiquitinated and D1 protein-containing nucleosomes within the Drosophila genome. Cell. 1982 Feb;28(2):375–385. doi: 10.1016/0092-8674(82)90355-5. [DOI] [PubMed] [Google Scholar]
- Matsui S. I., Seon B. K., Sandberg A. A. Disappearance of a structural chromatin protein A24 in mitosis: implications for molecular basis of chromatin condensation. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6386–6390. doi: 10.1073/pnas.76.12.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlhinney A., Hogan B. L. Rapid degradation of puromycyl peptides in hepatoma cells and reticulocytes. FEBS Lett. 1974 Apr 1;40(2):297–301. doi: 10.1016/0014-5793(74)80248-6. [DOI] [PubMed] [Google Scholar]
- Mueller R. D., Yasuda H., Hatch C. L., Bonner W. M., Bradbury E. M. Identification of ubiquitinated histones 2A and 2B in Physarum polycephalum. Disappearance of these proteins at metaphase and reappearance at anaphase. J Biol Chem. 1985 Apr 25;260(8):5147–5153. [PubMed] [Google Scholar]
- Ozkaynak E., Finley D., Varshavsky A. The yeast ubiquitin gene: head-to-tail repeats encoding a polyubiquitin precursor protein. Nature. 1984 Dec 13;312(5995):663–666. doi: 10.1038/312663a0. [DOI] [PubMed] [Google Scholar]
- Quail P. H., Briggs W. R. Irradiation-enhanced Phytochrome Pelletability: Requirement for Phosphorylative Energy in Vivo. Plant Physiol. 1978 Nov;62(5):773–778. doi: 10.1104/pp.62.5.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RABINOVITZ M., FISHER J. M. Formation of a ribosomal lesion in rabbit reticulocytes by the lysine antagonist, S-(beta-aminoethyl) cysteine. Biochem Biophys Res Commun. 1962 Jan 24;6:449–451. doi: 10.1016/0006-291x(62)90373-x. [DOI] [PubMed] [Google Scholar]
- Shanklin J., Jabben M., Vierstra R. D. Red light-induced formation of ubiquitin-phytochrome conjugates: Identification of possible intermediates of phytochrome degradation. Proc Natl Acad Sci U S A. 1987 Jan;84(2):359–363. doi: 10.1073/pnas.84.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegelman M., Bond M. W., Gallatin W. M., St John T., Smith H. T., Fried V. A., Weissman I. L. Cell surface molecule associated with lymphocyte homing is a ubiquitinated branched-chain glycoprotein. Science. 1986 Feb 21;231(4740):823–829. doi: 10.1126/science.3003913. [DOI] [PubMed] [Google Scholar]
- Solheim A. E., Seglen P. O. Structural and physical changes in lysosomes from isolated rat hepatocytes treated with methylamine. Biochim Biophys Acta. 1983 Oct 25;763(3):284–291. doi: 10.1016/0167-4889(83)90136-2. [DOI] [PubMed] [Google Scholar]
- Vierstra R. D., Langan S. M., Haas A. L. Purification and initial characterization of ubiquitin from the higher plant, Avena sativa. J Biol Chem. 1985 Oct 5;260(22):12015–12021. [PubMed] [Google Scholar]
- Vijay-Kumar S., Bugg C. E., Wilkinson K. D., Vierstra R. D., Hatfield P. M., Cook W. J. Comparison of the three-dimensional structures of human, yeast, and oat ubiquitin. J Biol Chem. 1987 May 5;262(13):6396–6399. [PubMed] [Google Scholar]
- Wharton S. A., Hipkiss A. R. Abnormal proteins of shortened length are preferentially degraded in the cytosol of cultured MRC5 fibroblasts. FEBS Lett. 1984 Mar 12;168(1):134–138. doi: 10.1016/0014-5793(84)80222-7. [DOI] [PubMed] [Google Scholar]
- Wheatley D. N., Henderson J. Y. P-fluorophenylalanine and 'division-related proteins'. Nature. 1974 Feb 1;247(5439):281–283. doi: 10.1038/247281a0. [DOI] [PubMed] [Google Scholar]