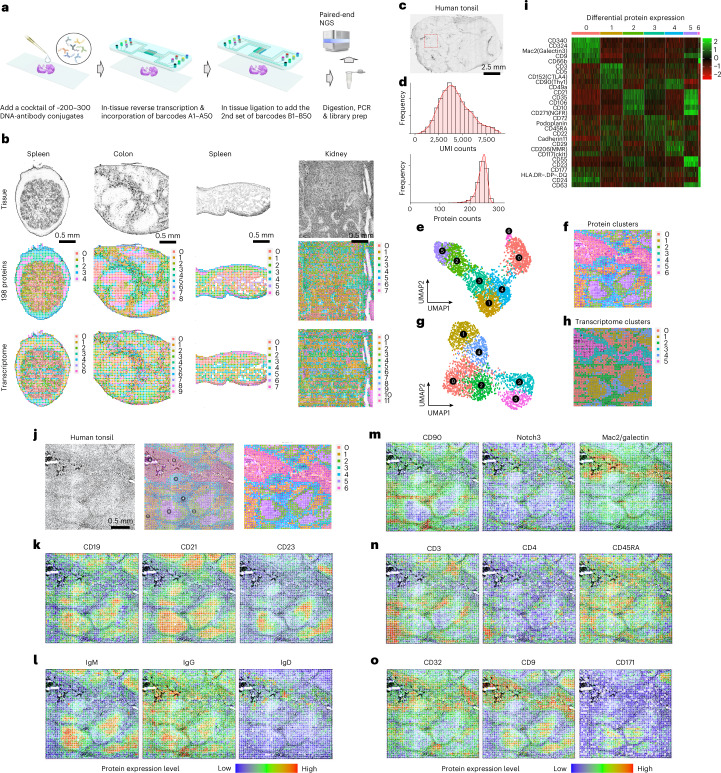
Fig. 1. Spatial-CITE-seq workflow design and application to diverse mouse tissue types and human tonsil for co-mapping of proteins and whole transcriptome.
a, Scheme of spatial-CITE-seq. A cocktail of ADTs is applied to a PFA-fixed tissue section to label a panel of ~200–300 protein markers in situ. Next, a set of DNA barcodes A1–A50 is flowed over the tissue surface in a spatially defined manner via parallel microchannels, and reverse transcription is carried out inside each channel for in-tissue synthesis of cDNAs complementary to endogenous mRNAs and introduced ADTs. Then, a set of DNA barcodes B1–B50 is introduced using another microfluidic device with microchannels perpendicular to the first flow direction and subsequently ligated to barcodes A1–A50, creating a 2D grid of tissue pixels, each of which has a unique spatial address code AB. Finally, barcoded cDNA is collected, purified, amplified and prepared for paired-end NGS sequencing. b, Spatially resolved 189-plex protein and whole transcriptome co-mapping of mouse spleen, colon, intestine and kidney tissue with 25-µm pixel size. Upper row: bright-field optical images of the tissue sections. Middle row: unsupervised clustering of all pixels based on all 189 protein markers only and projection onto the tissue images. Lower row: unsupervised clustering of whole transcriptome of all pixels and projection to the tissue images. Colors correspond to different proteomic or transcriptomic clusters indicated on the right side of each panel. c, Image of a human tonsil tissue section. The region mapped by spatial-CITE-seq is indicated by a dashed box. d, Per-pixel UMI count and protein count histograms. e, UMAP plot of the clustering analysis of all pixels based on 273 proteins only. f, Spatial distribution of the clusters (0–6) indicated by the same colors as in e. g, UMAP plot of the clustering analysis of all pixels based on the mRNA transcriptome. h, Spatial distribution of the transcriptomic clusters (0–5) indicated by the same colors as in g. Pixel size: 25 µm. i, Differentially expressed proteins in the clusters shown in c and d. j, Tissue image of the mapped region (left), spatial proteomic clusters (right) and the overlay (middle). k, Individual surface protein markers related to B cells and follicular DCs. l, Functional protein markers such as immunoglobulins showing spatially distinct distribution of GC B cells (IgM), matured B cells (IgG) and naive B cells (IgD), in agreement with B cell maturation, class switch and migration. m, Individual protein markers enriched in the extracellular region (CD90, Notch3) and crypt (Mac2). n, Individual T cell protein markers CD3, CD4 and CD45RA showing T cell zones and subtypes. o, Individual protein markers CD32, CD9 and CD171. CD32 identified a range of immune cells, including platelets, neutrophils, macrophages and DCs, trafficking from vasculature. CD9 identified plasma cell precursors in GCs and crypt. CD171, a neural cell adhesion molecule, is found highly distinct in the GC dark zone. Color key: protein expression from high to low.