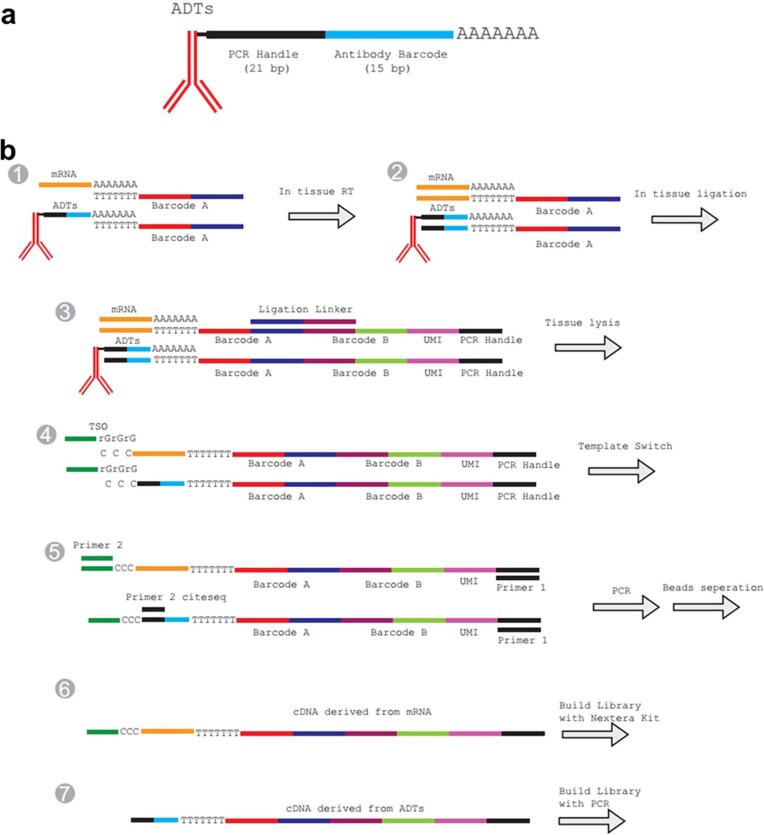
Extended Data Fig. 1. Spatial-CITE-seq design and detailed workflow.
(a) ADT structure. The oligo labelled to the antibody has three functional regions: PCR handle (21 bp), antibody barcode (15 bp) and poly-A region (32 bp). (b) ADTs and mRNA with Poly-A region at the 3′ end can be reverse transcribed into cDNA using Barcode A as the RT primer. Barcode A consists of three functional regions, the poly-T region, spatial barcode region and the ligation region. During the first flow, 50 Barcode As were loaded into 50 parallel channels and the RT reaction was carried out inside each isolated channel (Step 1&2). After peeling off the 1st PDMS, a 2nd PDMS was attached. The in-channel ligation was performed with injecting 50 Barcode Bs into each of the 50 channels which are perpendicular to the channels of 1st PDMS chip (Step 3). Barcode B has four functional regions: ligation region, barcode region, UMI region and PCR handle region. Barcode B was also 5′ biotin modification. After ligation, tissue was lysed, and cDNAs were purified with streptavidin beads. The cDNAs on the beads were templated switched with template switch oligo (Step 4). PCR was used to amplify the cDNA (Step 5). The products were split into two portions, the mRNA derived cDNAs and the ADT derived cDNAs. The library was then built separately. More details were in the method section.