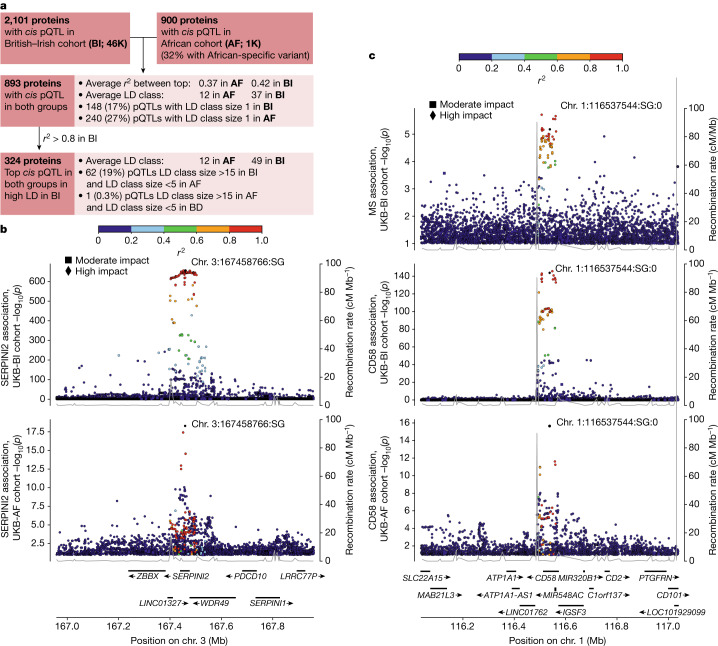
Fig. 2. Using different ancestry groups for locus refinement.
a, Using the more granular LD structure of the UKB-AF pQTLs to refine the location of pQTLs detected in the UKB-BI dataset. b, Locus plot of the sentinel cis pQTL for SERPINI2 in UKB-BI (top) and UKB-AF (bottom) ancestry groups. Although the sentinel cis pQTL for SERPINI2 is the same in the UKB-AF and UKB-BI groups, the LD class to which the variant belongs is much smaller in the UKB-AF group. This enables a more precise determination of which variant truly affects the protein levels. c, Locus plot of the association at the CD58 locus of the association with multiple sclerosis (MS) (top), and the sentinel cis pQTLs for CD58 in UKB-BI (middle) and UKB-AF (bottom). The locus refinement enabled by the smaller LD class in the UKB-AF group suggests that the disease association could be similarly refined. b,c, P values based on two-sided likelihood ratio test and not adjusted for multiple comparisons.