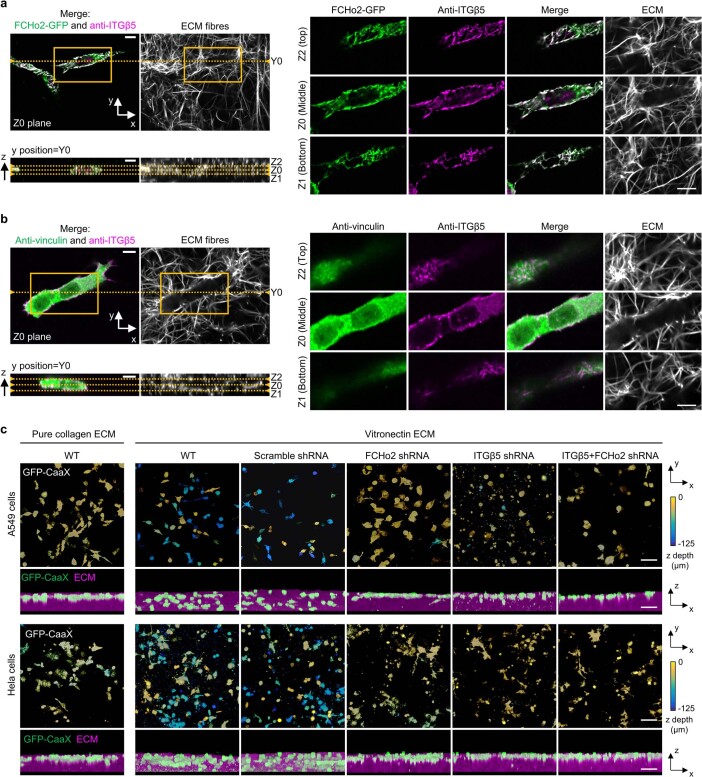
Extended Data Fig. 10. Curved adhesions in 3D ECMs facilitate cell migration in 3D ECMs.
a, x-y view (Z0 plane) and x-z view (Y0 plane) of z-stack images of immunolabelled ITGβ5 in U2OS cells expressing FCHo2-GFP. The cells are embedded in 3D ECM made of vitronectin fibres labelled with AF647-collagen. Zoom-in x-y images of the yellow-box area show abundant curved adhesions indicated by the colocalization of FCHo2 and ITGβ5. Curved adhesions form along vitronectin fibres at the middle (Z0 plane), bottom (Z1 plane), and top (Z2 plane) of cells. A zoom-in of the x-y slide at the Z0 plane has been shown in Fig. 5g (top). Scale: 10 µm. b, x-y view (Z0 plane) and x-z view (Y0 plane) of z-stack images of immunolabelled vinculin and ITGβ5 in U2OS. The cells are embedded in 3D ECM made of vitronectin fibres labelled with AF647-collagen. Zoom-in x-y images of the yellow-box area do not show clear focal adhesions indicated by the colocalization of vinculin and ITGβ5. Examination of different imaging planes, the middle (Z0 plane), bottom (Z1 plane), and top (Z2 plane) of cells, shows that the colocalization of vinculin and ITGβ5 is sparse. A zoom-in of the x-y slide at the Z0 plane has been shown in Fig. 5g (bottom). Scale: 10 µm. c, Representative 3D images of A549 cells (top) and HeLa cells (bottom) in 3D matrices after 72-hr culture. Cells were transduced to express GFP-CaaX via lentiviral infection. In the x-y projections, cells are colour-coded according to the z depth. In the x-z projections, cells are coloured in green and merged with the ECM (magenta). Cells can infiltrate into 3D matrices of vitronectin fibres, but not into 3D matrices of pure collagen fibres. The shRNA knockdown of FCHo2, ITGβ5, or both significantly inhibit cell infiltrations into 3D ECMs. Scale: 100 µm.