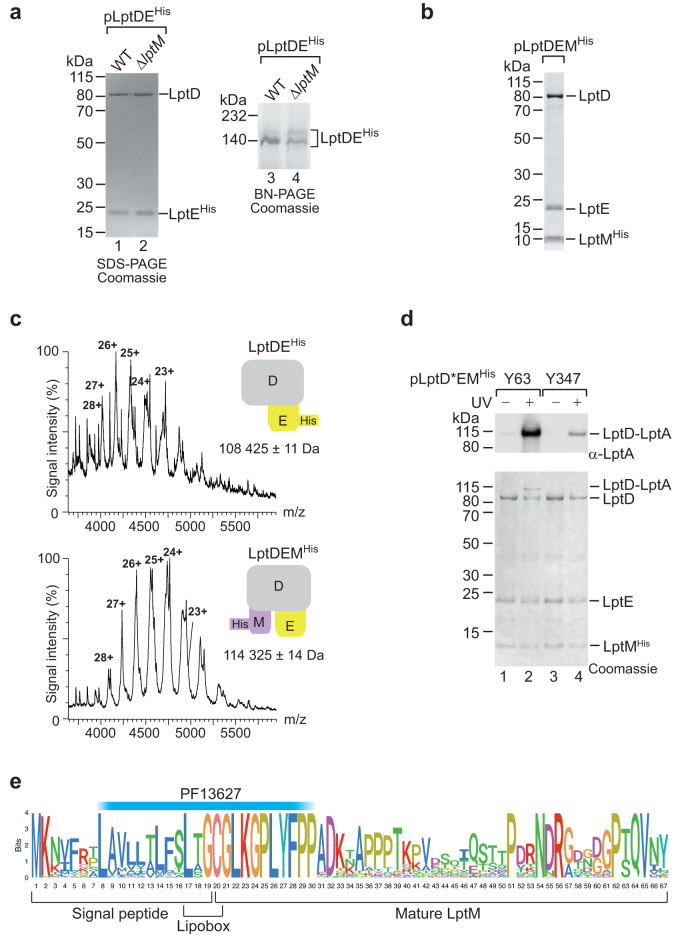
Fig. 1. LptM interacts with the LPS translocon.
a The envelope fractions of wild-type (WT, lptM+) and ΔlptM transformed with pLptDEHis cells were solubilized using the mild detergent DDM and subjected to Ni-affinity purification. The purified OM LPS translocon was analyzed by SDS-PAGE (lanes 1 and 2) and BN-PAGE (lanes 3 and 4) followed by Coomassie Brilliant Blue staining. Data are representative of three independent experiments. b DDM-solubilized LptMHis purified from ΔlptM cells transformed with pLptDEMHis was analyzed by SDS-PAGE and Coomassie Brilliant Blue staining. Data are representative of three independent experiments. c Native mass-spectrometry analysis of LptDEHis and of LptDEMHis purified as in (a) and (b), respectively. A schematic diagram of each complex is shown for each spectrum; LptD in gray, LptE in yellow, LptM in purple. Top: LptDEHis formed a heterodimeric complex of 108,425 ± 11 Da (LptD, theoretical mass of the mature protein = 87,080 Da; LptEHis, theoretical mass of the mature and tri-acylated protein = 21,348 Da). Bottom: LptDEMHis formed a heterotrimeric complex of 114,325 ± 14 Da (LptD, theoretical mass of the mature protein = 87,080 Da; LptE, theoretical mass of the mature and tri-acylated protein = 20,248 Da; LptMHis, theoretical mass of the mature and tri-acylated protein = 6,988 Da). The mass difference between the two complexes is 5,900 Da ± 25 Da, corresponding to the mass of mature tri-acylated LptM 5888 Da. d ΔlptM cells transformed with pLptDEMHis derivative (*) plasmids that carry pBpa at distinct positions in LptD were subjected to UV irradiation as indicated prior to DDM-solubilization and Ni-affinity purification of LptMHis. Upon SDS-PAGE of the elution fractions, proteins were revealed by Coomassie Brilliant Blue staining or Western blotting using an antiserum against LptA. Data are representative of three independent experiments. e Logoplot representation of the LptM amino acid sequence in Enterobacteriaceae. The plot was obtained from the multiple alignment of 37 amino acid sequences of LptM in representative genomes of a restricted Enterobacteriaceae family reported in Supplementary Fig. 4 (see also Supplementary Information). The location of PF13627 is indicated above the logoplot.