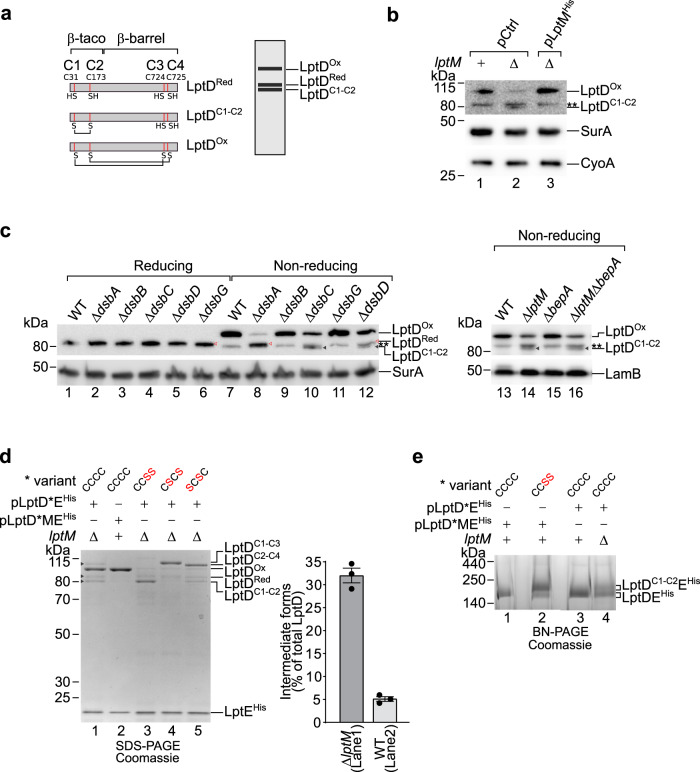
Fig. 3. LptM promotes LptD oxidative maturation.
a Schematic representation of mature LptD primary sequence indicating the position and oxidation state of Cys residues in LptDRed (top, left), LptDC1-C2 (middle, left) or fully oxidized LptDOx (bottom, left). A typical migration pattern corresponding to different oxidative states of LptD by non-reducing SDS-PAGE is represented on the right. b The total protein contents of wild-type (lptM+) and ΔlptM strains transformed with an empty vector pCtrl, and the complementation strain ΔlptM transformed with pLptMHis were separated by non-reducing SDS-PAGE and analyzed by Western blotting using the indicated antisera. **Indicates a non-identified protein band. Data are representative of three independent experiments. c The total protein contents of wild-type and the indicated mutant strains were separated by reducing or non-reducing SDS-PAGE and analyzed by Western blotting as in (b). Red, empty arrowheads indicate reduced LptD (LptDRed), whereas filled, gray arrowheads indicate LptDC1-C2. **Indicates a non-identified protein band that migrates slightly faster than LptDRed. Data are representative of three independent experiments. d Purification of LptEHis from ΔlptM cells that overproduced LptDEHis (lane 1) or from wild-type cells that overproduced LptDMEHis (lane 2) or ΔlptM cells that overproduced the indicated LptD mutant variant of LptDEHis (3-5). *Refers to the LptD Cys-to-Ser variant: wild-type LptDCCCC, LptDCCSS, LptDCSCS or LptDSCSC, as specified on the top of each gel lane. Data are representative of three independent experiments. The signals of any LptD forms in lanes 1 and 2 were quantified. The amount of intermediate forms (LptDRed + LptDC1-C2 + LptDC2-C4) was normalized to the overall amount of all LptD forms (LptDRed + LptDC1-C2 + LptDC2-C4 + LptDox). Data are represented as means ± s.e.m. (n = 3 independent experiments). Source data are provided as Source Data file. e LptDEHis containing either wild-type (LptDCCCC) or LptDCCSS co-overproduced together with LptM or alone in wild-type or ΔlptM cells were Ni-affinity purified and resolved by BN-PAGE, prior to Coomassie Brilliant Blue staining. *Indicates the plasmid-encoded LptD variant, wild-type LptDCCCC, LptDCCSS, as specified on the top of each gel lane. Data are representative of three independent experiments.