Figure 3. Benchmarking of MERCI for real-world application.
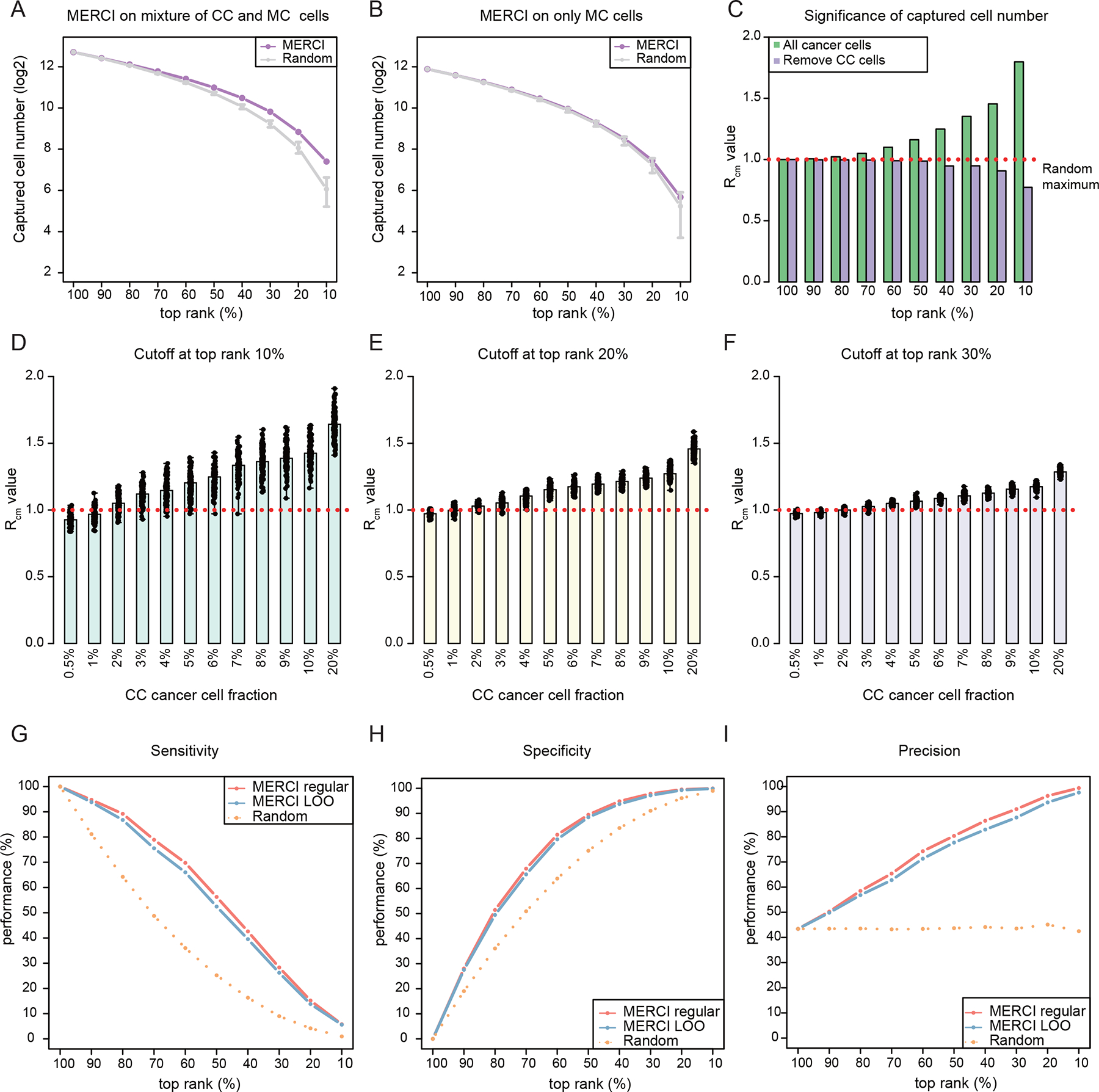
(A-B) Dot-line plots showing the number of positive calls captured by using AND gate to MERCI DNA and RNA rank scores across a range of cutoffs. Purple dots represent the results of in silico mixture sample and gray intervals indicate the ranges established by 10,000 randomly permutated ranks. Error bars indicate the range (min to max) of the data. (C) Barplots showing the values at different rank cutoffs. Red dotted line indicates . (D-F) Significance estimation of positive calls when different fractions of true receivers are included. Barplots showing the averaged values reported by MERCI for down-sampled datasets at rank cutoffs at top 10% (D), 20% (E) and 30% (F) respectively. Black dots indicate the values of down-sampled datasets. (G-I) The sensitivity, specificity and precision of MERCI when using different rank cutoffs to predict the MT receivers. See also Figure S3.
