Figure 4. Cycling gene expression can result from various kinetic modes that often involve dynamically regulated RNA decay.
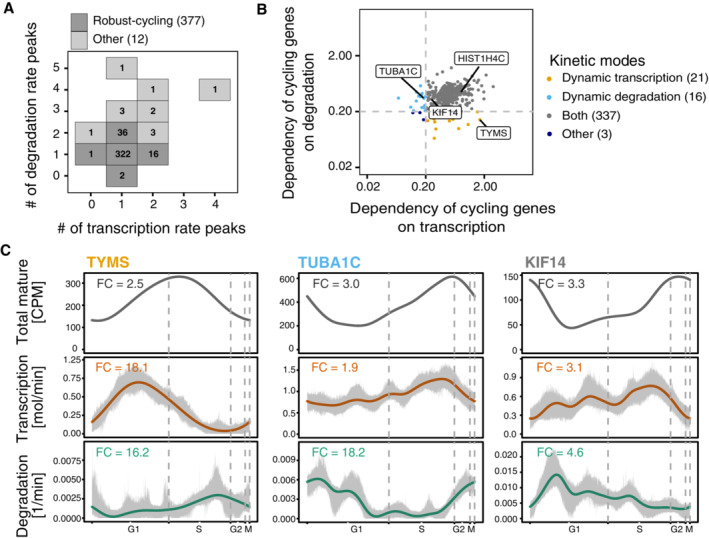
- Identification of the robust‐cycling genes for kinetic rate regulation investigation. 377 out of the 389 well‐predicted cycling genes were defined as robust‐cycling genes by excluding those showing multiple transcription or degradation peaks.
- Cycling genes exhibit diverse kinetic modes. The kinetic modes were defined based on the dependency of dynamic transcription and dynamic degradation. The dependency on transcription (or degradation) is represented by the mean relative deviation between prediction from constant transcription (or degradation) and prediction from dynamic model (see Materials and Methods). ‘Both’ means genes are regulated by both dynamic transcription and dynamic degradation. Genes that were not identified into dynamic transcription, dynamic degradation or both were assigned into the “other” group. The labeled genes represent example cases in each mode.
- Profiles of the predicted expression, transcription and degradation rates along the cell cycle for example genes, which are labeled in (B) (dynamic transcription: TYMS; dynamic degradation: TUBA1C; dynamic transcription and dynamic degradation: KIF14). Gray areas around rates' profiles indicate the 90% confidence intervals calculated from bootstrapping (resampling of cells 100 times with replacement).
