Figure 1. APOE and CLU RNA and protein levels are reduced in SORL1 KO neurons.
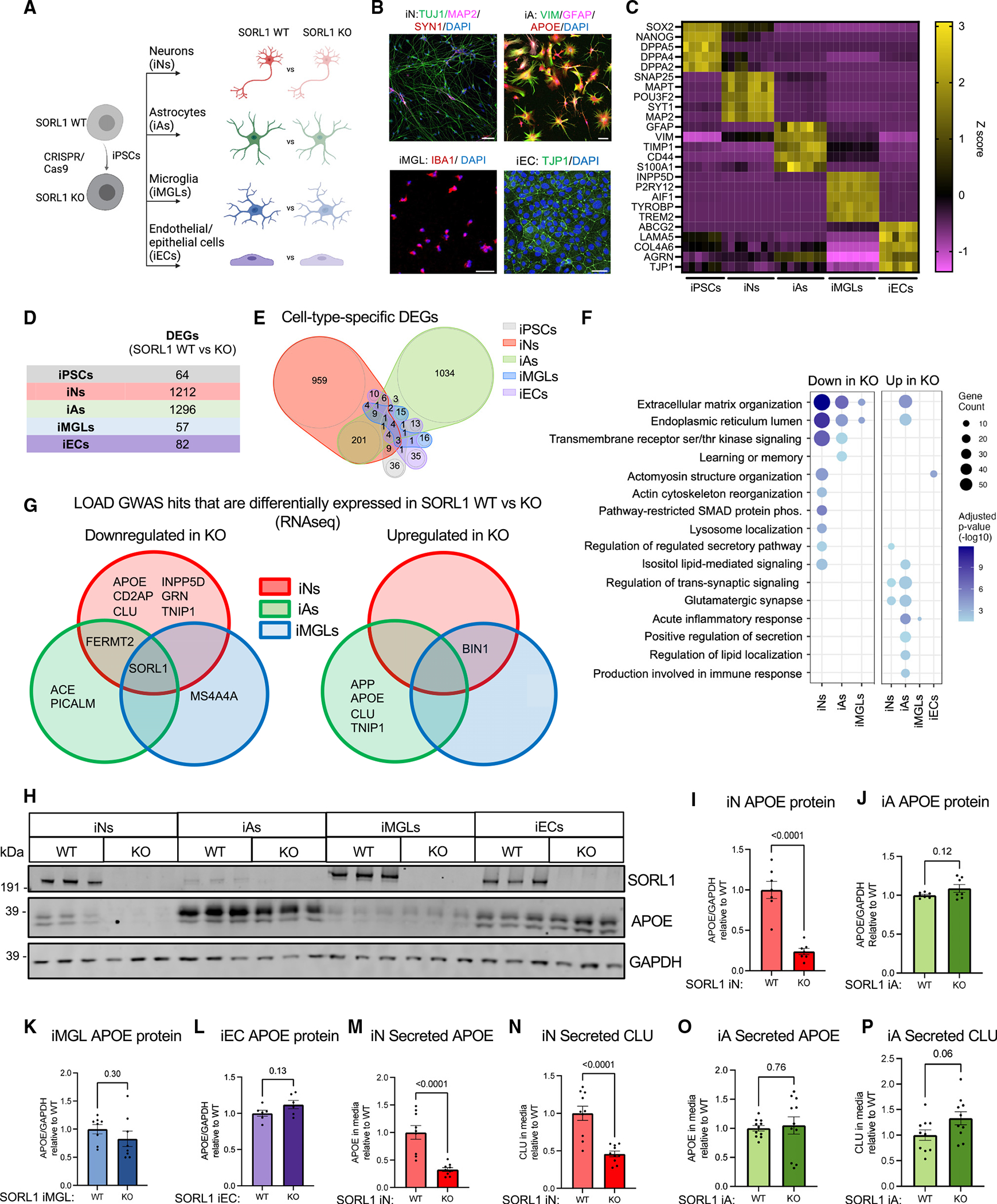
(A) Schematic of the experimental design. SORL1 KO iPSCs were generated using CRISPR-Cas9 as previously published,16 and paired KO and WT iPSCs were differentiated into neurons (iNs), astrocytes (iAs), microglia (iMGLs), and endothelial/epithelial cell (iEC) fates.
(B) Representative immunocytochemistry images of iN, iA, iMGL, and iEC cultures. Scale bars, 50 μm.
(C) Heatmap of RNA expression (Z score) of cell-type-specific markers.
(D and E) Cultures were analyzed via RNA-seq and differential expression analyses performed within each cell type. DEGs were identified using Sleuth40 by performing a Wald test (false discovery rate [FDR] q < 0.05 and b < −0.5 or b > 0.5). The numbers of DEGs between SORL1 WT and KO in each cell type are shown as a table (D) and as a Venn diagram (E). Complete datasets can be found in Tables S1.
(F) Dot plot showing significantly overrepresented Gene Ontology (GO) pathways in iNs, iAs, iMGLs, and iECs comparing SORL1 KO and WT within each cell type. Dots are sized based on the number of DEGs in the indicated pathway and colored based on adjusted p value.
(G) Venn diagram of LOAD GWAS genes that are differentially expressed in SORL1 KO neurons, astrocytes, and microglia. Genes with adjusted p <0.05 are included in the diagram.
(H) Representative western blot of iNs, iAs, iMGLs, and iECs showing protein expression levels of SORL1, APOE, and GAPDH.
(I–L) Quantification of APOE/GAPDH in iNs, iAs, iMGLs, and iECs.
(M–P) Secreted APOE and CLU values were measured using MSD ELISA, and the values were normalized to WT within each comparison. For each panel, three independent differentiations were performed. In each round of differentiation, three or four wells were included for each group. Comparisons only performed within cell type; paired Student’s t test (two tailed), p values listed.
