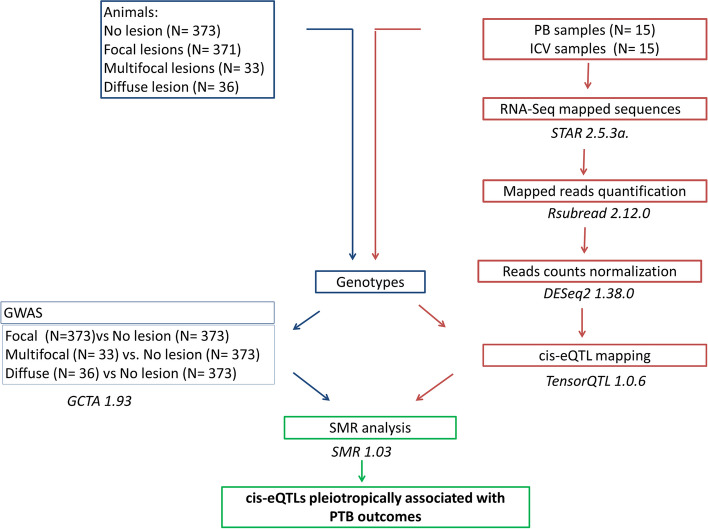
Fig. 1.
Study design. Summary data from GWAS and cis-eQTL mapping were used to perform the SMR analysis. The approach starts with the individual normalized RNA-counts, then identifies genetic variants controlling differences in gene expression, and finally checks whether the identified cis-eQTLs are indeed associated with disease outcomes using summary GWAS data from a bigger population classified according to the presence or absence of PTB-associated lesions. Two eQTL datasets from peripheral blood (PB) and ileocecal valve (ICV) were applied together with three GWAS datasets from the comparisons: focal, multifocal or diffuse lesions versus (vs) controls. Using these two different omics data, the SMR method discover biological mechanisms behind PTB outcomes