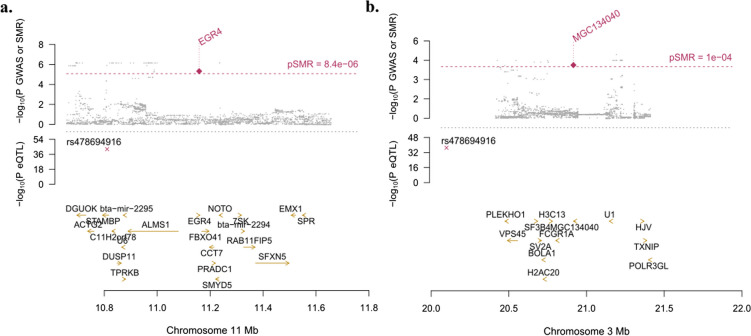
Fig. 3.
Locus plots showing the significant genes EGR4 and MGC134040, their locations within chromosomes 11 and 3, respectively. Y axis represents the negative log of the significant P-values and X axis represents the negative log of the P-values. Dots in the top plots represent the P-values for SNPs from the GWAS analysis. Rhombuses represent the P-values from the SMR analyses, highlighted in red if significant. In the middle plots, crosses represent the Pβ-values of the cis-eQTLs located in the plotted region. In the bottom plots are represented all genes in each plotted region. a. SMR results using cis-eQTLs identified in PB samples and GWAs results of a case–control study where cases were animals with multifocal lesions and controls were animals without lesions. The dotted red line represents FDR ≤ 0.05 corresponding to P [SMR] = 10–5. b. SMR results using ci-eQTLs identified in ICV samples and GWAs results of a case–control study where cases were animals with diffuse lesions and controls were animals without lesions. The dotted red line represents FDR ≤ 0.05 corresponding to P [SMR] = 10–4