Fig. 2 |. BigMHC network architecture and pseudosequence composition.
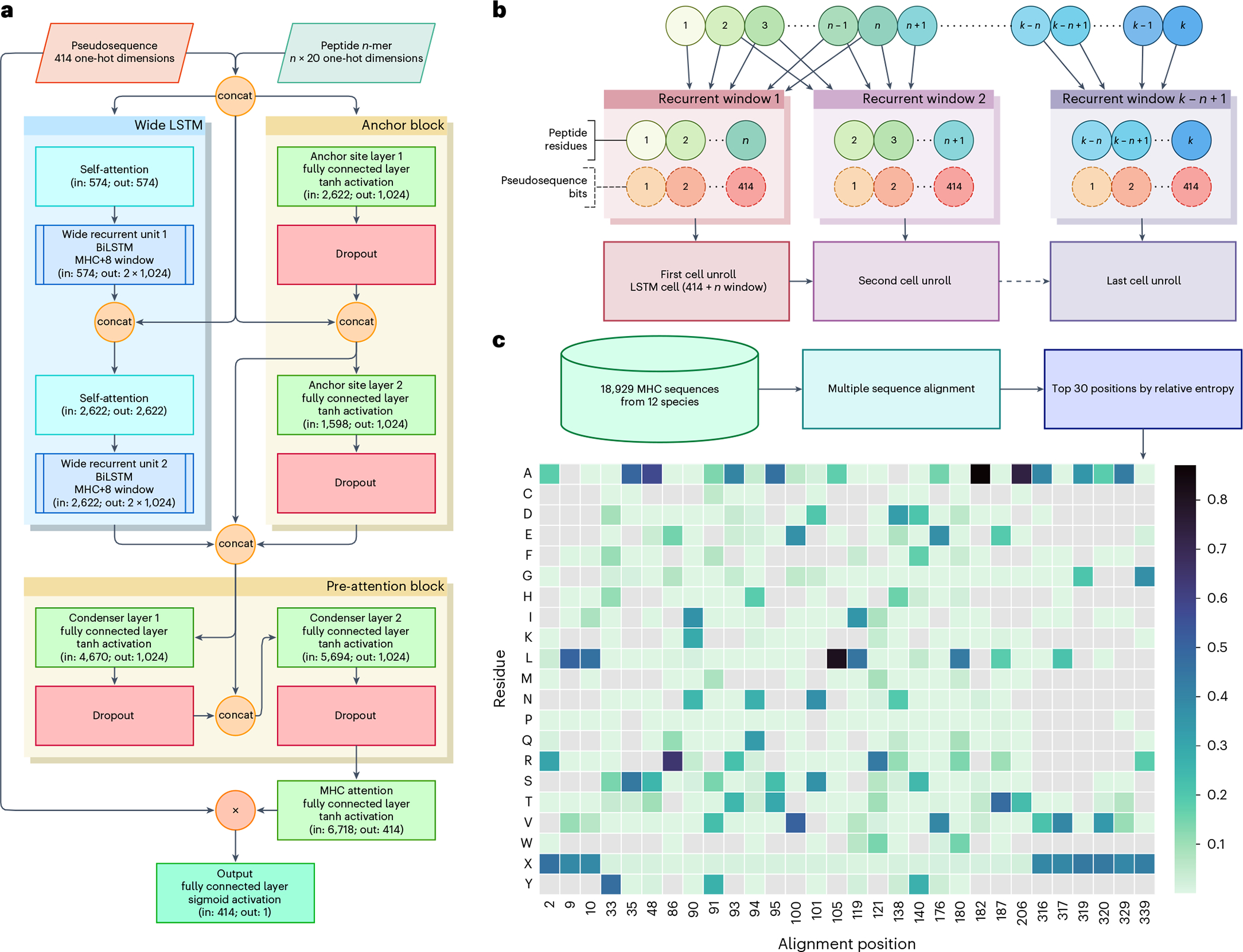
a, The BigMHC deep neural network architecture, where the BigMHC ensemble comprises seven such networks. Pseudosequences and peptides are one-hot encoded prior to feeding them into the model. The circles labelled ‘Con’ indicate concatenation and the circle labelled ‘×’ denotes element-wise multiplication. The anchor block consists of two densely connected layers that each receive the first and last four peptide residues along with the MHC encoding. The self-attention modules are single-headed attention units, which is analogous to setting the number of heads of a standard multi-headed transformer attention module to one. Prior to the final sigmoid activation, the output of the model is a weighted sum of the MHC pseudosequence one-hot encoding; the weights are referred to as attention. Because all connections except internal BiLSTM cell connections are dense, data are not bottlenecked until the MHC attention node maps the pre-attention block output to a tensor of the same shape as the one-hot-encoded MHC pseudosequences. b, A wide LSTM. Each cell unroll processes the entire MHC pseudosequence but only a fixed-length window of the peptide. Where a canonical LSTM uses a window length of one, BigMHC uses a window length of eight to capitalize on the minimum pMHC peptide length. c, The pseudosequence amino acid residue probability (represented by the color scale) per alignment position. Note that not all amino acid residues are present for each position, as indicated by grey cells, so the one-hot encoding uses a ragged array, encoding only the residues present at a given position.
