Abstract
The cellular complexity of the human brain is established via dynamic changes in gene expression throughout development that is mediated, in part, by the spatiotemporal activity of cis-regulatory elements (CREs). We simultaneously profiled gene expression and chromatin accessibility in 45,549 cortical nuclei across six broad developmental time points from fetus to adult. We identified cell type–specific domains in which chromatin accessibility is highly correlated with gene expression. Differentiation pseudotime trajectory analysis indicates that chromatin accessibility at CREs precedes transcription and that dynamic changes in chromatin structure play a critical role in neuronal lineage commitment. In addition, we mapped cell type–specific and temporally specific genetic loci implicated in neuropsychiatric traits, including schizophrenia and bipolar disorder. Together, our results describe the complex regulation of cell composition at critical stages in lineage determination and shed light on the impact of spatiotemporal alterations in gene expression on neuropsychiatric disease.
Single cell analysis of the developing brain unravels cell fate determination and the molecular basis of neuropsychiatric disease.
INTRODUCTION
Human brain development starts during the early stages of embryogenesis and extends postnatally through infancy, childhood, adolescence, and young adulthood (1, 2). To produce distinct circuits in the human cortex, neurons are born in an immature state and undergo a variety of molecular and morphological changes as they differentiate, migrate, and establish synaptic networks. Environmental and genetic risk factors can disrupt these highly orchestrated developmental processes, potentially leading to neuropsychiatric disease (3, 4). Given the variable age of onset of different neurodevelopmental disorders, it is critical to examine the effect of risk factors across the full spectrum of human brain development.
The developmental transition of cell lineages is highly orchestrated by dynamic changes in gene expression, mediated in part by spatiotemporal patterns of transcription factor (TF) binding to cis-regulatory DNA elements (5–9). Over the past decade, there has been an extensive effort to explore changes in the transcriptome and, to some extent, epigenome in the developing brain (10). Initial studies on bulk tissue samples surveyed broad spatiotemporal trajectories of gene expression (5) as well as dynamics of underlying molecular regulators (11). Single-cell transcriptome analysis has expanded our knowledge of cellular diversity and the molecular changes that occur during differentiation, migration, and synaptic network formation in the human cortex (6, 9, 12–15). Although single-cell transcriptomics have been the primary focus of research, a recent study (16) also performed epigenetic profiling to examine the gene expression regulatory mechanisms. However, there has been no simultaneous multi-omic (joint gene expression and chromatin accessibility) single-cell profiling in the developing brain. The multi-omic profiling has emerged as a means to decipher how combinations of TFs drive gene expression programs and to infer cell lineage transitions during development (17). Consequently, joint analysis of gene expression and chromatin accessibility at the single-cell level can provide a more complete understanding of the gene-regulatory dynamics associated with human brain development.
To that end, we generated a transcriptomic and chromatin accessibility data collection, profiling 45,549 cells using multi-omic single-nucleus RNA sequencing (snRNA-seq) and assay for transposase-accessible chromatin with sequencing (ATAC-seq), across a broad developmental time frame that includes human fetal cortical plate, early postnatal, adolescent, and adult specimens. We explored gene-regulatory interactions by combining chromatin accessibility with gene expression within the same cells, and identified a subset of genes that are regulated by multiple nearby putative enhancers and have an important role in lineage determination during cortical development. To better understand the regulatory mechanisms driving neurogenesis, we performed pseudotime trajectory analysis and detected dynamic changes in chromatin accessibility preceding transcript production as a critical component of neuronal lineage commitment. We evaluated the enrichment of lineage-specific genes and chromatin accessible regions with genetic risk loci for neuropsychiatric disorders to explore their cellular ontogeny. Together, our data present a valuable resource for understanding the gene-regulatory dynamics associated with human brain development and for prioritizing targets for further study as well as the generation of therapeutics to treat neurodevelopmental disorders.
RESULTS
Single-nucleus gene expression and chromatin accessibility profiles revealed congruent cell types in the human cortex
We used the 10X Chromium Single Cell Multiome ATAC + Gene Expression kit to simultaneously profile the transcriptome (via snRNA-seq) and chromatin accessibility (via snATAC-seq) in 12 samples of human neocortex from six developmental periods (early mid gestation fetal, late mid gestation fetal, infancy, childhood, adolescence, and adulthood) (Fig. 1A and table S1). To confirm that the paired profiles were truly derived from the same cells, we first performed multi-omic profiling on two samples containing mixtures of human and mouse cell lines, and asked whether the coassayed cells were consistently assigned to the same species labels. As expected, no doublets were identified and we observed that human and mouse reads were well separated on the basis of the chromatin and transcriptome profiles of filtered cells (fig. S1A).
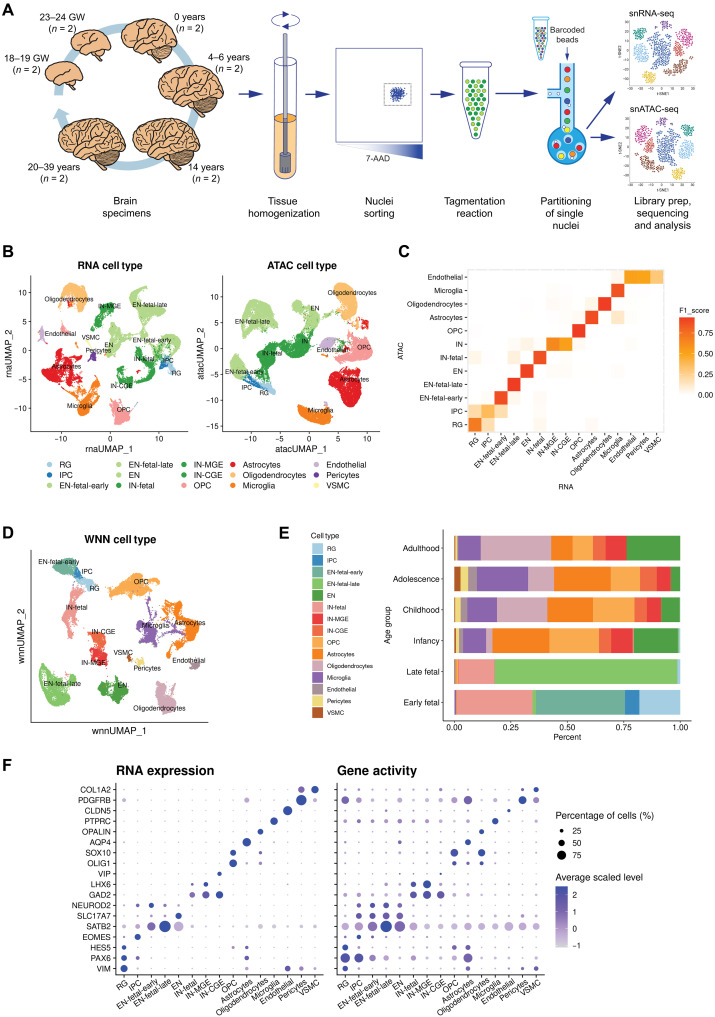
Fig. 1. Joint single-cell profiling of RNA expression and chromatin accessibility of human neocortex.
(A) Frozen human cortical brain specimens from six developmental time points were homogenized and purified by FANS before tagmentation and partitioning into single nuclei using the 10x Genomics platform. Libraries for snRNA-seq and snATAC-seq were prepared, sequenced, and analyzed independently. (B) UMAP visualizations of single cells defined by RNA-seq and ATAC-seq data, respectively. Cell type annotations are derived from either modality independently. (C) Heatmap showing the concordance of cell memberships between the two clustering results, measured in F1 score. (D) UMAP visualization of single cells defined by integrating two modalities using WNN analysis. Cell type annotations are determined on the basis of marker genes. (E) Proportions of cell types in each age group. (F) Dot plot showing selected marker gene expression and chromatin-derived gene activity across cell types.
We then processed human neocortex samples, obtaining joint profiles of chromatin accessibility and gene expression from 45,549 of 53,185 single nuclei that met quality control and filtering criteria (Materials and Methods). To assess the similarities and differences between the two modalities, we first clustered the RNA-seq and ATAC-seq datasets independently (Fig. 1B and Materials and Methods). Broadly, both modalities revealed the same major neocortical cell types and that cell identities assigned to RNA-seq– and ATAC-seq–derived cell types were highly congruent [Fig. 1C; adjusted Rand index (ARI) = 0.78].
Similar to previous multi-omic single-cell studies (17, 18), the independent modality analyses exhibit differences, primarily in the composition of cell populations in the fetal and postnatal stages (fig. S1B). On one hand, some cell types were broadly identified but not distinguishable in the ATAC-seq clustering results. For example, the medial ganglionic eminence (MGE)–derived and caudal ganglionic eminence (CGE)–derived inhibitory neuron subtypes were not distinguished; various stromal cell types with smaller population sizes, including endothelial cells, pericytes, and vascular smooth muscle cells (VSMCs), were blended together. On the other hand, RNA-seq data showed insufficient power to identify progenitor cells, as evidenced by nearly 20% fewer detected radial glia (RG) and intermediate progenitor cells (IPCs) when compared with the ATAC-seq results (1427 for RNA-seq versus 1743 for ATAC-seq), indicating that active gene-regulatory dynamics at different developmental stages might be better reflected in chromatin accessibility than in the transcriptome (19). These results motivated us to anticipate more comprehensive information about cell type classifications by leveraging both modalities.
Joint analysis of multi-omic data improves cell type identification
We next performed joint clustering on the paired modalities of the same single cells using a weighted-nearest neighbor (WNN) analysis (18). WNN is an unsupervised method that generates an integrated representation of cellular identity by learning the information content of each modality. The WNN analysis results were in agreement with those derived from either single modality (ARI = 0.88 for RNA-seq, ARI = 0.86 for ATAC-seq), while the inferred relative modality weights varied across cell types (fig. S1C), reflecting the biological importance of each modality in determining cellular identity. The WNN analysis resulted in 28 clusters, including all the major and minor cell types in the human brain cortex, which were further grouped into 15 cell types (Fig. 1D and Materials and Methods). We confirmed that each cluster comprised cells from different samples (fig. S1D), suggesting that taxonomy was not determined by donor or other technical covariates.
Gene activity inferred by gene expression and chromatin accessibility of known cell type–specific markers consistently confirmed cluster identity (Fig. 1F and Materials and Methods). Specifically, we found neural progenitor cells (NPCs) expressing PAX6, including RG (one cluster; HES5 and VIM) and IPCs (one cluster; EOMES). We also identified three subtypes of excitatory neurons (SATB2, SLC17A7, and NEUROD2) representing different developmental stages, one enriched for cells from early fetal samples (“EN-fetal-early”; four clusters), one for late fetal samples (“EN-fetal-late”; two clusters), and the third for postnatal samples (“EN”; two clusters). Similarly, there were three subtypes of inhibitory neurons identified (GAD1 and GAD2), two of which represent MGE-derived (“IN-MGE”; one cluster; LHX6) and CGE-derived (“IN-CGE”; one cluster; VIP and ADARB2) subtypes in postnatal samples, while the remaining subtype was enriched in fetal samples (“IN-fetal”; one cluster). The types of neurons that are distinct between fetal and postnatal human brain samples support previous findings (20). In addition, we observed clusters of major glial cell types in the neocortex, including oligodendrocyte progenitor cells (OPCs; two clusters; OLIG1 and SOX10), astrocytes (three clusters; AQP4 and GFAP), oligodendrocytes (three clusters; MOBP and OPALIN), microglia (four clusters; PTPRC and CX3CR1), as well as endothelial cells (one cluster; CLDN5), pericytes (one cluster; PDGFRB), and VSMCs (one cluster; COL1A2) (see the lists of differentially expressed genes and accessible peaks in tables S2 and S3).
Sample-specific cell type composition varied markedly across developmental stages (Fig. 1E). In the four fetal samples, neuronal populations accounted for the vast majority of cells, whereas postnatal samples had much higher proportions of nonneuronal cells. The changing patterns of cell type composition were in line with the results from a previous deconvolution study using multiple bulk and single-cell datasets (21). Moreover, we found that most of the neural progenitors (91%), including the transient cell types of RG and IPCs, were only detected in the two early fetal samples [gestational week (GW) 18 to 19; Fig. 1E and fig. S1E], consistent with the fact that the bulk of neurogenesis in the human cerebral cortex has occurred by midgestation (at GW20) and these progenitor cells start disappearing or transforming with the completion of cortical development (9, 22). Notably, the results derived from joint analysis identified every cell type that was found in either single-omic analyses while not losing power for detection of neural progenitors (1736 by joint analysis versus 1743 by ATAC-seq alone versus 1427 by RNA-seq alone).
Cis-regulatory associations between chromatin peaks and target genes revealed extensive regulatory interactions
Multi-omic data offer the advantage to explore gene-regulatory interactions by combining chromatin accessibility with gene expression within the same cells. Because of the sparsity of snATAC-seq and snRNA-seq data, we examined the relationships between the two modalities using pseudobulk aggregates rather than individual cells (17, 19, 23). We generated 500 pseudobulk samples by aggregating RNA-seq and ATAC-seq signals from similar cell types (fig. S2A and Materials and Methods).
First, we sought to globally quantify the relative contribution of proximal (i.e., promoter) and distal (i.e., enhancer) chromatin accessibility to transcriptional variance. We applied a variance component model to the expression of each gene using the covariance of chromatin accessibility at promoter and enhancer regions as inputs, and corrected for donor and age effects by adding the inter-individual and inter–age group covariance to the model (Materials and Methods) (24, 25). This approach does not model the relationship of each gene to its own promoters or enhancers, but instead models the genome-wide relationships to all enhancers or promoters. Our results suggested that more than 80% of expression variance was attributed to promoter and enhancer accessibility (Fig. 2A), indicating that transcriptional heterogeneity is broadly associated with the variation of chromatin accessibility. As a control experiment, we randomly permuted the dataset and, as expected, a minimal proportion of variance (<1%) was explained by the epigenome in the shuffled analysis (fig. S2B). There was a small group of genes (n = 56) for which >60% of the expression variance could be best explained by the inter–age group covariance. Gene ontology (GO) enrichment analysis of these genes revealed enrichment in DNA binding transcriptional activators [false discovery rate (FDR) q value = 0.02; including those that encode known TFs such as SOX11, SOX4, NEUROD6, and NR3C1], suggesting the temporal role for these TFs in human brain development.
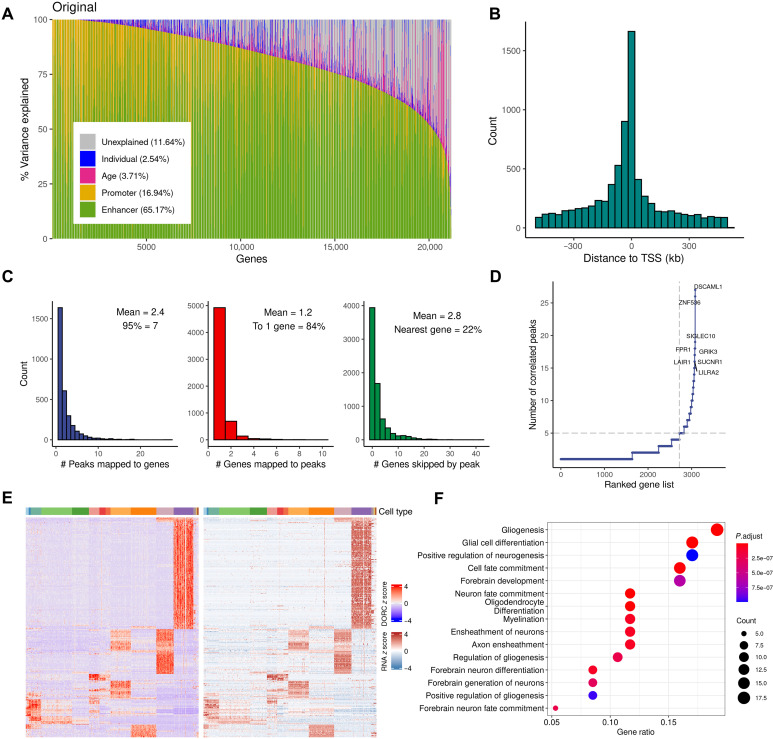
Fig. 2. Global and local characterization of cis regulation patterns.
(A) Variance component analysis showing chromatin accessibility explains variation in gene expression. Genes, in columns, are sorted by the decreasing proportion of variance explained by the epigenome (enhancers and promoters), with the mean-variance explained by each component shown in parenthesis. (B) Distribution of the distance from each peak to the TSS of the linked gene. (C) Histograms showing (from left to right) distribution of the number of peaks significantly linked per gene; distribution of the number genes significantly linked per peak; distribution of the number of genes “skipped” by a peak to reach its linked gene. (D) Number of significantly linked peaks for each gene, with genes sorted in increasing order. (E) Heatmap showing chromatin accessibility and gene expression of the linked peak-gene pairs (rows, left: aggregated peak accessibility, right: linked gene expression) in the DORCs across 500 pseudobulk samples (columns, sorted in terms of cell types); values are z score normalized. (F) Top 15 GO enrichment results for genes linked to DORCs.
With the aim of linking a regulatory element to its target gene(s), we next used a correlation-based approach to examine the pairwise relationships between chromatin accessibility and gene expression (Materials and Methods). This led to the identification of 7291 significant peak-gene associations [within ±500 kb around transcription start sites (TSSs), Spearman correlation coefficient |ρ| > 0.3, FDR-adjusted P < 0.1; table S4], involving 3082 unique genes. The majority (97.6%) of these links included peaks that were positively correlated with gene expression (fig. S2C). As expected, these associations were enriched in the vicinity of TSSs, and the correlations decayed exponentially with distance (Fig. 2B). Nevertheless, only 22% of the peak-gene links occur between an ATAC-seq peak and the nearest gene, indicating that most predicted regulatory interactions skip at least one gene along the linear genome (Fig. 2C), demonstrating the shortcomings of purely applying the “nearest neighbor gene” rule to define regulatory targets (24, 26, 27). The expression of most genes is, on average, correlated with at least two different peaks, while most peaks (84%) are predicted to interact with a single target gene (Fig. 2C). To validate the set of identified peak-gene links, we used the “activity-by-contact” (ABC) approach (28) (Materials and Methods) and compared them with the enhancer-promoter (E-P) interactions that were previously derived from the matched bulk brain tissues (26). We observed significantly higher ABC scores in the group of E-P interactions overlapping with the peak-gene links (P < 2.2 × 10−16 by Wilcoxon test; fig. S2D), thereby providing further validation.
Cell type–specific cis-regulatory domains determine cell lineage during cortical development
To investigate the specificity of peak-gene associations across cell types and developmental stages, we assigned each interaction to the cell type with the highest average gene expression and chromatin accessibility. Peak-gene associations were strongest in the early developmental stages, while they became diminished in more differentiated stages (fig. S2E). Specifically, RG-specific peak-gene links were the strongest across all cell types; in the group of neurons (either excitatory or inhibitory), which consists of samples from fetal to postnatal stages, we observed a clear weakening pattern of the associated links with developmental age. We defined a “pseudo-age” for each cell type (Materials and Methods) and confirmed a significantly negative relationship with the median link strengths (Pearson’s r = −0.57, P = 0.026; fig. S2E).
Despite the fact that most genes involved in peak-gene links were associated with one or two peaks, a subset of genes were associated with a relatively large number of peaks, suggesting orchestrated coregulation of the target gene activity by multiple factors that act upon a broad chromatin domain. In total, we identified 364 domains of regulatory chromatin (DORCs) (17) in which there are at least five significant peak-gene links associated with the same gene (Fig. 2D and Materials and Methods). In previous studies, it has been shown that DORCs are often associated with super-enhancers—large clusters of enhancer regions that are known to play key regulatory roles in defining cell identity and are affected across multiple diseases (29, 30). Consistent with these studies, we found that DORCs identified here were also prominently overlapped with super-enhancers, which were identified by using neuronal and glial chromatin immunoprecipitation sequencing (ChIP-seq) H3K27 acetylation data from human brain samples (P = 8.3 × 10−68 by hypergeometric test; table S5) (26). For example, the DORC of the DSCAML1 gene contained 27 peak-gene associations. The epigenetic dysfunction of this super-enhancer has been implicated in Alzheimer’s disease (AD) pathology (31).
Motivated by a previous study (17), we hypothesized that DORCs are highly cell type specific. We defined a DORC score for each gene as the aggregated normalized counts from all peaks significantly associated with that gene (Materials and Methods). Covariation of chromatin accessibility and gene expression distinguished the identified cell types in both RNA-seq and ATAC-seq data (Fig. 2E), suggesting the cell type specificity of DORC-gene links. GO analysis of the genes involved in the top decile of the peak-gene correlations in DORCs revealed strong enrichment of developmental processes in both neurons and glia (Fig. 2F), highlighting the important role of DORCs in cell fate determination during cortical development. Through comparison of neurons from different developmental stages (table S6), we found a higher number of DORCs specific to earlier stages (e.g., fetal versus postnatal and early fetal versus late fetal), suggesting a role in regulating early neurodevelopmental processes.
Chromatin priming precedes gene expression during neuronal lineage commitment
Having identified various neuronal subtypes from early fetal cortical plate to adult cortical samples, we next used the paired multi-omic single nuclei profiles to infer the developmental dynamics of gene regulation throughout corticogenesis and neuron differentiation. We performed a pseudotime trajectory analysis by focusing on the neuronal populations (including RG, IPC, EN-fetal-early, EN-fetal-late, EN, IN-fetal, IN-MGE, and IN-CGE) and by anchoring the starting point in the RG cluster (Materials and Methods). Different cell types were properly laid on the inferred trajectories in terms of their developmental stages (Fig. 3A and fig. S3A), with the fetal sample–specific neuronal populations located between the initial progenitor populations and the mature neurons from postnatal samples (i.e., EN, IN-MGE, and IN-CGE). The developmental trajectories separated into EN lineage and IN lineages shortly after the starting point, and the IN lineage later split into IN-MGE and IN-CGE subtypes (Fig. 3B). The respective numbers of cells assigned to each of the three lineages are as follows: EN lineage (14,146), IN-MGE lineage (5728), and IN-CGE lineage (4904).
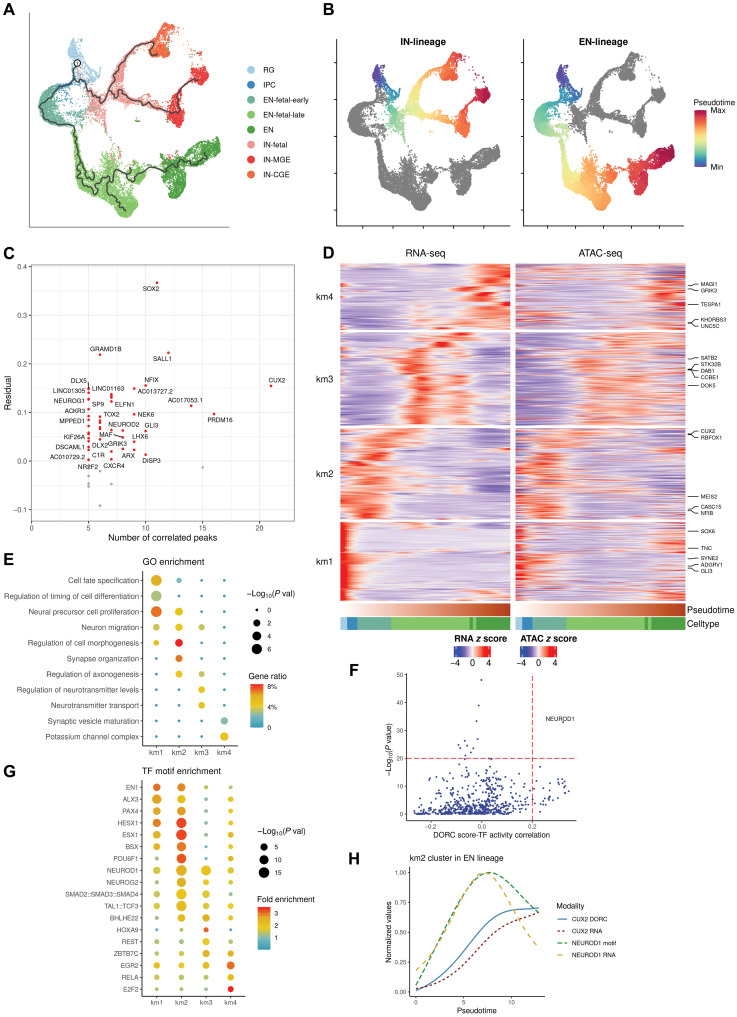
Fig. 3. Trajectories of gene regulation during neuronal development.
(A) Trajectories identified within the neuronal subpopulations, shown on the RNA gene expression coordinates (root node was annotated as “1”; cells were colored for annotated cell types). (B) Inferred pseudotime along the lineages for excitatory neurons (“EN-lineage”) and inhibitory neurons (“IN-lineage”), respectively. (C) Average residuals between chromatin accessibility and gene expression versus the number of significantly linked peaks for each gene involved in the DORCs identified within the neuronal populations. Positive and negative residuals are colored in red and gray, respectively. (D) Heatmap showing gene expression and DORC chromatin accessibility of the peak-gene links that significantly varied along the pseudotime for the EN lineage. Rows (genes) are clustered using k-means clustering (k = 4), and columns (cells) are ordered by pseudotime. The top five most differentially expressed genes in each cluster (km1/2/3/4) are annotated. (E) Respective GO enrichment of genes represented in the four peak-gene link clusters of the EN lineage. (F) P values of TF motif enrichment in km2 peaks plotted against Spearman correlation of TF motif activity with CUX2 DORC score. (G) TF motif enrichment of peaks represented in the peak-gene link clusters of the EN lineage. (H) Lineage dynamics of NEUROD1 motif activity and expression precede CUX2 DORC chromatin accessibility and gene expression in the EN lineage, from the beginning to the end of the km2 stage, using the min-max normalized, smoothed values over pseudotime.
We repeated the peak-gene association analysis focusing on the neuronal populations (Materials and Methods), resulting in 1638 significant associations involving 930 unique genes (table S7). Similarly, we defined 55 neuron-specific DORCs (associated with at least five peaks), which strongly overlapped with the DORCs that we defined using all cells (P = 5.8 × 10−51, by hypergeometric test). GO analysis of the genes involved in these DORCs revealed a significant enrichment for neuron differentiation pathways as well as the overrepresentation of DNA binding TFs, either activators or repressors (table S8).
Given the potentially tight regulation of DORC target genes by dynamic changes in chromatin accessibility during lineage commitment, we sought to explore whether chromatin accessibility at DORCs precedes gene expression. For each of the DORCs, we quantified a “residual” by subtracting the corresponding gene expression value from the DORC score (Materials and Methods) (17). We observed that the residuals were typically positive (46 of 55) across lineages (Fig. 3C), which reflected the lineage priming of cis-regulatory elements, as the DORCs generally became accessible before onset of their associated gene’s expression. Furthermore, we found that the lineage-priming pattern became more robust for DORCs with a higher number of peaks, indicating higher confidence in the chromatin accessibility–primed states. Overall, these findings suggest that dynamic changes in chromatin accessibility are a critical component of neuronal lineage commitment, similar to previous observations during hair follicle differentiation (17).
NEUROD1 induces CUX2 chromatin priming and gene expression during development of excitatory neurons
We next looked deeper into the peak-gene links on the EN lineage, which started from neuronal progenitors, including RGs and IPCs, and then differentiated into excitatory neuron subtypes specific to different developmental stages sequentially, from early fetal to late fetal and then to postnatal (Fig. 3, A and B). We found that the expression levels of over 87% of the linked genes (811 of 930) varied significantly along the pseudotime trajectory (Materials and Methods). We then grouped these genes into four clusters using k-means (km) clustering, each of which corresponding to a different developmental period (Fig. 3D). GO enrichment analysis on this gene set revealed the unique biological activities occurring during different time periods (Fig. 3E). Specifically, at the beginning of the trajectory (“km1”), the linked genes were enriched in processes relating to cell fate specification, timing regulation of cell differentiation, and neural precursor cell proliferation. In the next early fetal period (km2), the peak-gene interactions became associated with neuron migration, morphogenesis, synapse organization, and axonogenesis. Afterward, in the late fetal (km3) and postnatal stages (km4), the excitatory neurons acquired the ability for neurotransmitter transport and regulation, indicating cell maturation.
The dynamic regulatory activities during the developmental transition of cell lineages is highly orchestrated by the spatiotemporal patterning of TFs. To identify TFs that control these dynamic regulatory activities, we performed TF motif enrichment analysis in the different clusters (Materials and Methods). TF motifs with an established function in cell differentiation and development were enriched in the earliest stage, including for EN1, which has been implicated as a crucial mediator of dopaminergic subset specification (32), and HESX1, which is encoded by a hub gene involved in neural commitment (Fig. 3G and table S9) (33). In the intermediate stages (including early and late fetal), the associated peak-gene links were more enriched in motifs of neuronal TFs such as NEUROD1, NEUROG2, and BHLHE22, suggesting that the most active neurogenesis processes occur during these particular developmental periods. Fewer TF motifs were found enriched in the last postnatal peak-gene link cluster, including cell cycle regulators such as E2F2.
Cut-like homeobox 2 (CUX2) was identified as a neuron-specific DORC gene, regulated by the highest number (n = 21) of nearby putative enhancers (Fig. 3C), and as a marker for the second earliest stage in the EN lineage (km2; Fig. 3D). This is consistent with the well-known function of CUX2 as a neuron-specific TF regulating dendritic branching and synapse formation (34). Next, we investigated which TF(s) might activate CUX2 enhancers by leveraging the correlation between the DORC score of CUX2, the TF motif activity (inferred from ATAC-seq), and the TF motif enrichment for the km2 cluster (Materials and Methods). The binding motif for the TF NEUROD1 was strongly enriched in km2 chromatin accessible regions, and NEUROD1 activity was highly correlated with the CUX2 DORC chromatin accessibility state (Fig. 3F). NEUROD1 is essential for eliciting the neuronal development program and has the ability to reprogram other cell types into neurons (35). We next ordered single cells based on the inferred pseudotime for the km2 stage and identified a clear pattern where the activity of NEUROD1 precedes the CUX2 DORC chromatin state, followed by CUX2 gene expression (Fig. 3H). In addition, we found that, as NEUROD1 activity decreases, the rate of CUX2 expression slows down accordingly. These results suggest that NEUROD1 is likely a key TF during early neurogenesis (35) to induce CUX2 DORC accessibility followed by CUX2 transcription.
Repression of NEUROD1 expression in cultured NPCs suppresses CUX2 expression
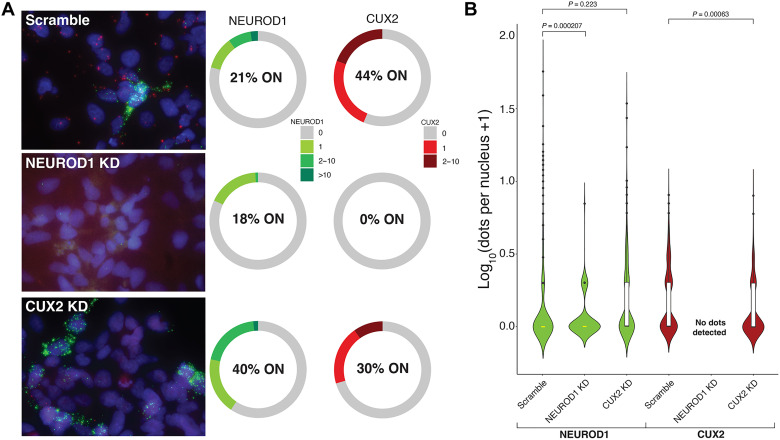
We sought to validate the predicted causal relationship between NEUROD1 and CUX2 by performing CRISPRi in cultured NPCs followed by RNAscope to directly image mRNAs in single cells. We transfected guide RNAs (gRNAs) against both NEUROD1 and CUX2 into NPCs stably expressing dCas9-KRAB and then differentiated the NPCs using an established protocol (36). In the negative control experiment, in which cells were treated with a scrambled gRNA, we observe that CUX2 is widely expressed, albeit at low numbers per cell, whereas NEUROD1 expression is restricted to a smaller subset of cells but with a broader range of mRNA numbers per cell, including a fraction displaying strong bursts of transcription (Fig. 4A). We quantified the frequency distribution of fluorescent dots per nucleus for both genes at week 2 after differentiation and found that NEUROD1 expression is more variable than CUX2 across the population, as measured by the fano factor [(variance/mean), CUX2 = 1.78, NEUROD1 = 17.02] (Fig. 4B).
Fig. 4. Assessment of the relationship between NEUROD1 and CUX2 in differentiating NPCs.
(A) Maximum intensity projection images (from 200-nm z stacks obtained at ×63 magnification) of CUX2 (red) and NEUROD1 (green) expression in NPCs 2 weeks after differentiation treated with scrambled gRNA, a NEUROD1-specific gRNA, and a CUX2-specific gRNA. Charts show frequency distributions of RNAscope dots per nucleus for CUX2 and NEUROD1 in cells treated with scrambled gRNA (n = 444 cells), NEUROD1-specific gRNA (n = 111 cells), and CUX2-specific gRNA (n = 183 cells). % ON corresponds to % of nuclei with detectable RNAs. (B) Violin plots of nuclear RNA frequency distributions in all conditions. A two-sided Wilcoxon rank sum test with continuity correction was performed. The center line (yellow) indicates the median, the box shows the interquartile range, and whiskers indicate the highest/lowest values within 1.5× the interquartile range.
Inactivation of NEUROD1 led to a down-regulation of NEUROD1 mRNA compared to the control (P = 0.0002 by negative binomial test), while CUX2 transcription is completely suppressed (Fig. 4, A and B). Given that around 80% of nuclei in the control NPCs do not show transcription of NEUROD1, the NEUROD1 promoter may be tightly repressed for long periods but allows for infrequent, strong bursts of transcription. In turn, inactivation of CUX2 with CRISPRi led to down-regulation of CUX2 mRNA and a decrease in the proportion of cells expressing CUX2 compared to control (P = 0.0006 by negative binomial test). Although we only achieved partial suppression of CUX2, this led to an apparent (albeit insignificant, P = 0.223 by negative binomial test) increase in expression of NEUROD1, suggesting that CUX2 may either directly or indirectly regulate NEUROD1, an observation that is consistent with previous studies implicating CUX2 as a negative regulator of gene expression (37), including in cortical neurons (34, 38). Together, our data suggest that although NEUROD1 is expressed infrequently, it is required to maintain ongoing transcription of CUX2.
Leveraging cell type–specific genes and peaks in heritability analysis allows dissociation of risk loci for neuropsychiatric traits
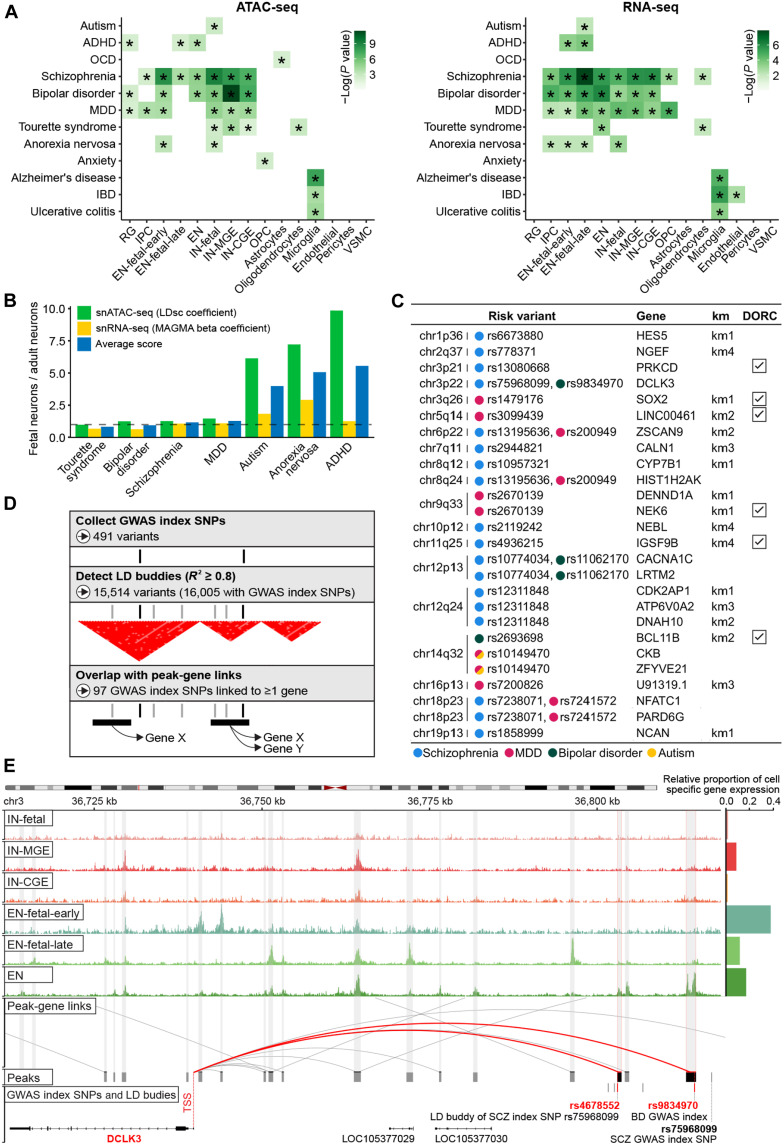
Despite the notable progress in exploring the genetic causes of neuropsychiatric disorders, their underlying molecular mechanisms are still not fully understood (39). To reveal whether disorder-associated variants are enriched in a particular cell type or developmental stage, we used linkage disequilibrium (LD)–aware approaches (Materials and Methods) (40, 41) to assess the overlap between a collection of relevant genome-wide association studies (GWASs) and lineage-defining genes and chromatin peaks derived from our multi-omic single-cell data. By analyzing 9 neuropsychiatric and 3 unrelated control traits in 15 cell types, we identified 33 and 28 significant associations in cell type–specific chromatin accessibility and transcriptome data, respectively (Fig. 5A and tables S10 and S11). We observed a high overlap of significant cell type—GWAS trait pairs (20 of 41 of significant pairs are shared; Spearman correlation of all pairs ρ = 0.62), suggesting that both modalities report reliable and informative associations.
Fig. 5. Mapping of risk variants associated with neuropsychiatric traits to causal genes using single cell–derived marker genes and peaks.
(A) Heritability enrichment of brain cell types in neuropsychiatric disorders and unrelated control traits. Heatmaps highlight significant colocalization of GWAS-derived common genetic variants with cell-specific open chromatin regions in snATAC-seq data (left) and cell marker genes in snRNA-seq data (right) (Materials and Methods). “*”: significant after correction across all tests (FDR < 0.05). (B) Comparison between fetal and adult neuronal signals in selected neuropsychiatric disorders (traits need to be enriched in either fetal or adult category; therefore, OCD and anxiety were not involved). Fetal and adult neurons are represented by peak sets/gene sets compiled from unions of the top 2500/500 most cell-specific peaks/genes from each fetal neuron (i.e., EN-fetal-early, EN-fetal-late, and IN-fetal) and adult neuron (i.e., EN, IN-CGE, and IN-MGE) category. To calculate the ratio “fetal neurons/adult neurons” (y axis), we used LDsc regression coefficients (snATAC-seq) and MAGMA beta coefficients (snRNA-seq); joint score is an average of snATAC-seq and snRNA-seq scores. (C) Subset of candidate causal genes for risk variants that either are prioritized in two disorders or show significantly altered expression along the developmental trajectory of the neuronal lineage (km1/2/3/4; full list of causal genes in table S12). (D) Schematic of the overall strategy to connect risk variants associated with neuropsychiatric disorders to their causal genes (Materials and Methods). (E) Normalized snATAC-seq–derived pseudobulk tracks demonstrating the complex cell-specific regulation of the DCLK3 gene that is predicted to be the causal gene for SCZ and BD GWAS risk variants (rs75968099 and rs75968099).
Consistent with previous studies, schizophrenia (SCZ), bipolar disorder (BD), attention deficit hyperactivity disorder (ADHD), and major depressive disorder (MDD) are enriched in multiple neuronal subtypes (42, 43). However, to our knowledge, our results identified several associations that have not yet been described by genetic data (see Discussion), including oligodendrocytes for Tourette syndrome (TS), astrocytes for obsessive-compulsive disorder (OCD), OPCs for anxiety, and fetal neurons for anorexia nervosa (AN). For nonpsychiatric, immune-related traits, including AD, ulcerative colitis, and inflammatory bowel disease, we only observed enrichment for microglia, thus further strengthening the credibility of our results. To dissect the temporal specificity in neuropsychiatric diseases, we compared fetal and adult neuronal enrichment using both epigenome and transcriptome data. We found a high concordance between enrichment of both assays, allowing us to classify ADHD, AN, and autism as being more strongly associated with fetal neuronal stages, while, for TS, BD, SCZ, and MDD, we found an equal contribution of both fetal and adult neuronal stages (Fig. 5B). Analysis of rare exome (44, 45) and copy number variants (46, 47) for SCZ and autism identified similar cell types (fig. S4), further supporting the mechanistic convergence of common and rare risk variants across neurodevelopmental disorders. In addition, de novo disruptive and benign variants identified by whole-genome sequencing of autism cases and controls were enriched with DORCs and epigenome markers (Materials and Methods and table S13) associated with differentiation of late fetal excitatory neurons (table S13), suggesting the highly negative impact of mutations within regulatory regions with important roles in development.
We next aimed to nominate the candidate functional genes for disease-associated loci (Fig. 5D and Materials and Methods). First, we collected a set of 491 genome-wide significant variants associated with neuropsychiatric traits (P < 5 × 10−8) and extended it to 16,005 variants based on the presence of high LD (R2 ≥ 0.8). We overlapped putative disease-relevant variants [index single-nucleotide polymorphisms (SNPs) and LD buddies] with the peaks that demonstrate significant peak-gene associations to pinpoint at least one gene under regulation for 97 genome-wide significant loci (table S12). Of 152 genes mapped to those 97 loci, 7 were linked to two disease traits simultaneously and 17 genes were shown to have significantly altered expression along the pseudotime trajectory of neuronal lineage specification (km1/2/3/4) (Fig. 5C). While the original GWAS usually nominate several plausible gene targets for each disease-relevant locus, their prioritization is mostly based on imprecise distance-based annotation. Using our approach, we were able to refine their predictions and, in some cases, nominate target genes not previously associated with the disease. One example of a replicated finding is the association of DCLK3 (encoding a neuroprotective kinase) with both SCZ and BD, which was previously observed by transcriptome-wide association study (TWAS) (48) and expression quantitative trait loci (eQTL) approaches (Fig. 5E) (49). Notably, this association is derived from the overlap of putative disease-relevant variants with two distinct peaks, both predominantly accessible in adult excitatory neurons.
DISCUSSION
We generated multimodal chromatin accessibility and gene expression data in the human cortex across six broad developmental time points from fetus to adult. Joint analysis of 45,549 individual nuclei facilitated the identification of genes and cis-regulatory elements with fundamental roles in lineage determination. Our results are consistent with a recent study implicating chromatin accessibility as a predetermining factor of lineage commitment in murine skin cells (17). By using the covariance of chromatin accessibility at promoter and enhancer regions as inputs, we show that most expression variance was attributed to promoter and enhancer accessibility, indicating that gene expression is broadly associated with chromatin accessibility. Similar results have been reported in human blood cells following immune stimulation (50). Moreover, through comparison of neurons from different developmental stages, we found that there were more DORCs specific to earlier stages (e.g., fetal versus postnatal and early fetal versus late fetal), suggesting a role for chromatin reorganization in regulating early neurodevelopmental processes.
The temporal nature of our data allowed us to examine neural trajectories across four broad developmental phases. The first of these contains genes involved in cell fate specification, differentiation, and NPC proliferation. The second cluster specifies genes involved in neuron migration, morphogenesis, synapse organization, and axonogenesis, while the third and fourth clusters contain genes associated with neurotransmitter transport and regulation. As an example, we chose to focus on CUX2, a TF involved in synaptogenesis that is expressed in the second cluster. CUX2 expression coincides with a number of nearby open chromatin regions containing putative enhancers, among which are binding sites for NEUROD1, a well-known pioneer factor involved in neuronal cell fate specification. It has recently been shown that overexpressing NEUROD1 in astrocytes can convert them into neurons, suppressing the astroglial gene expression program while up-regulating neuronal genes, including CUX2 (51). Thus, we hypothesized that NEUROD1 might activate CUX2 during early neural development and subsequently showed that the inactivation of NEUROD1 in cultured NPCs led to a complete suppression of CUX2.
In recent years, large-scale GWAS have identified numerous, broadly defined loci associated with increased risk for a range of brain-related phenotypes. Concurrent technological advances, in particular, those that facilitate examination of tissues at the single-cell level, have, in turn, significantly improved our understanding of the genetic basis of brain development and function (6, 9, 12–15). Previous single-cell studies, however, focused on separate modalities performed independently. By assaying the epigenome and gene expression in parallel in individual cells, we sought to better understand the role played by the regulome during brain development and to more accurately pinpoint causative risk variants within GWAS loci. Lineage-specific genes and chromatin accessible regions are enriched for risk loci associated with neuropsychiatric traits and implicate 152 putative risk genes in a range of disorders, including SCZ, BD, ADHD, and MDD. SCZ, BD, ADHD, and MDD are enriched in multiple neuronal subtypes, consistent with previous studies (16, 42–43). The spatiotemporal resolution of our data allowed us to categorize common risk variants from ADHD, AN, and autism as more strongly associated with fetal neuronal stages compared to TS, BD, SCZ, and MDD. Despite the fact that rare variants are thought to contribute only marginally toward explaining missing heritability and population-level risk (52), we found concordant patterns of enrichment in cell type and developmental age between rare and common risk variants for autism and SCZ, suggesting that they are at least partially mechanistically convergent (44, 52, 53).
Beyond already known associations between various cell types and diseases, our results identified several associations that, to our knowledge, have not been described previously. First, TS was found to be enriched in oligodendrocyte cells in both epigenome and transcriptome assays. The critical role of oligodendrocytes is supported by tract-based spatial statistics measurements of TS patients, indicating a reduced fractional anisotropy that reflects deficits in axonal myelination (54). Second, OCD was enriched in astrocytes. While the literature supporting this relationship is more established for the striatum (55), the involvement of the prefrontal cortex was previously studied through an astrocyte-specific deletion of glutamate transporter 1 (56), resulting in OCD-like behavior. Third, anxiety was enriched in OPCs, further implicating the well-established role for aberrant myelination in neuropsychiatric disorders (57). This relationship was emphasized by a recent study linking anxiety-like behavior in a mouse model of cuprizone-induced demyelination, which displays impaired OPC differentiation (58). Last, we report the enrichment of fetal neurons in AN. While this disorder phenotypically manifests in adolescence or early adulthood, a number of studies suggest substantial changes during earlier stages of development (59, 60). Furthermore, significant differences in gene expression were previously measured between AN case and control subjects using human induced pluripotent stem cell (hiPSC)–derived cortical neurons (61), which are known to resemble fetal, rather than adult, brain cells (62).
In conclusion, we generated a dataset of gene expression and chromatin accessibility in single nuclei from six developmental time points that provides additional insights into cell fate determination in the human cerebral cortex and on the molecular basis of neuropsychiatric disease. We present our data as an interactive web browser that can be used by the scientific community to explore spatiotemporal alterations in gene expression in development and disease.
MATERIALS AND METHODS
Description of postmortem brain samples
Brain samples for this study were selected on the basis of age (six age groups) and sex (one male and one female in each age group). Fetal brain samples were collected from deidentified prenatal autopsy specimens without neuropathological abnormalities at the Icahn School of Medicine at Mount Sinai. The cortical plate was dissected fresh from the anterior frontal lobe of anatomically intact brain specimens. Young and old brain samples were accessed through the National Institutes of Health NeuroBioBanks at the University of Miami Brain Endowment Bank and Mount Sinai Brain Bank, respectively. All neuropsychological, diagnostic, and autopsy protocols were approved by the respective Institutional Review Boards. No sample size calculation was performed. For further processing, we randomized brain samples for sex and age.
Isolation and FANS
All buffers were supplemented with ribonuclease inhibitors (Takara). Four samples were processed in parallel. Twenty-five milligrams of frozen postmortem human brain tissue was homogenized in cold lysis buffer [0.32 M sucrose, 5 mM CaCl2, 3 mM magnesium acetate, 0.1 mM EDTA, 10 mM tris-HCl (pH 8), 1 mM dithiothreitol (DTT), and 0.1% Triton X-100] and filtered through a 40-μm cell strainer. The flow-through was underlaid with sucrose solution [1.8 M sucrose, 3 mM magnesium acetate, 1 mM DTT, and 10 mM tris-HCl (pH 8)] and centrifuged at 107,000g for 1 hour at 4°C. Pellets were resuspended in phosphate-buffered saline (PBS) supplemented with 0.5% bovine serum albumin (BSA). Before fluorescence-activated nuclear sorting (FANS), volumes were brought up to 250 μl with PBS and 7-Amino-Actinomycin D (7AAD) (Invitrogen) was added according to the manufacturer’s instructions. 7AAD-positive nuclei were sorted into tubes precoated with 5% BSA using a FACSAria flow cytometer (BD Biosciences).
Multiome ATAC-seq and gene expression library preparation
Following FANS, nuclei were subjected to two washes in 200 μl of wash buffer (10x Genomics), after which they were resuspended in 30 μl of nuclei buffer (10x Genomics) and quantified (Countess II, Life Technologies). Eight thousand nuclei from each sample were subjected to the Chromium Next GEM Single-Cell Multiome ATAC-seq and Gene Expression protocol (10x Genomics) according to the manufacturer’s instructions. Resulting libraries were quantified using the KAPA library quantification kit (KAPA Biosystems), and fragment sizes were determined by Tapestation (Agilent). All libraries were sequenced at New York Genome Center using the Novaseq platform (Illumina), obtaining 100–base pair (bp) paired-end reads. Individuals generating libraries were not blinded. We did not exclude any preselected brain sample or generated library. We generated one library for each brain sample.
Data processing and quality control
Fastq alignment, filtering, barcode counting, peak calling, and counting of both ATAC-seq and gene expression molecules were performed with cellranger-arc (v.1.0.0). Then, we applied Seurat v4.0 (18) and Signac v1.1.0 (63) to create a multi-omic Seurat object with paired gene expression and DNA accessibility profiles for each sample. For chromatin accessibility, we used MACS2 (64) as implemented in the function CallPeaks in Signac to call peaks from the fragment files. We removed any peaks on nonstandard chromosomes overlapping annotated genomic blacklist regions from the hg38 genome (65). We then quantified fragment counts for each peak, per cell, using the FeatureMatrix function in Signac. Per-cell quality control metrics for each modality were computed, including mitochondrial percentage, nucleosome signal, and TSS enrichment score. Next, we combined the individual Seurat objects for all the samples into one single object using the merge function in Seurat. We then performed quality control based on metrics for both modalities by retaining cells with total RNA-seq count >200 and <50,000, total ATAC-seq count >200 and <100,000, mitochondrial percentage <5%, nucleosome signal <3, and TSS enrichment score >1. Genes or peaks that were detected in <10 cells were removed. As a result, 45,549 of 53,185 single cells were retained, with 5887 RNA Unique Molecular Identifier (UMI) counts and 18,066 unique ATAC-seq fragments (29.3% fragments overlapping peaks) on average. We applied Scrublet (66) for doublet detection and the results showed that less than 1% of the total cell population were suspected as doublets, and the numbers for different cell types are listed in table S14. Given the small proportion, we did not exclude these cells from the analysis. We tested that all samples showed at least moderate TSS enrichment (fig. S1F) and followed a stereotypical distribution of fragment lengths with a majority of short fragments (under 100 bp) followed by a tail of longer fragments (over 147 bp) in multiples of the nucleosomal unit size (fig. S1G).
We normalized the gene expression count data using SCTransform (67) and then performed principal components analysis (PCA) using the RunPCA function in Seurat. The first 30 principal components (PCs) were used in the downstream analysis. For the chromatin accessibility data, we performed latent semantic indexing (LSI) for dimension reduction (68). We first normalized the ATAC-seq peaks using the log-TF version of the term frequency–inverse document frequency (TF-IDF) transformation by using the RunTFIDF function in Signac (parameter setting: “method = 3”). The top 25% most frequently observed peaks of the TF-IDF matrix were then selected by the FindTopFeatures function for singular value decomposition (SVD) using the RunSVD function in Signac. The top 10 LSI components, excluding the first, were used in downstream analysis. The first LSI component was excluded because it typically captures sequencing depth (technical variation), as evidenced by the high correlation with the total number of counts for the cells (>0.5). We also created a gene activity matrix inferred from ATAC-seq by using the GeneActivity function in Signac, which assesses chromatin accessibility at gene body and promoter regions.
Clustering and visualization
We performed graph-based clustering using the reduced dimensions that we selected for both assays, as described above. For joint multi-omic analysis, we used the function FindMultiModalNeighbors in Seurat v4.0 to construct a weighted nearest neighbor (WNN) graph by taking as input two-dimensional reductions computed for each modality (18). WNN identifies the nearest neighbors for each cell based on a weighted combination of two modalities. For single modality assay, we used the function FindNeighbor to construct a shared nearest neighbor graph. We then applied the Smart Local Moving algorithm (69) on the derived graphs by using the function FindClusters (parameter settings: “algorithm = 3” and “resolution = 0.2”).
For two-dimensional visualization, we performed Uniform Manifold Approximation and Projection (UMAP) (70) as implemented in the function RunUMAP in Seurat on the PCs and LSI components for gene expression and DNA accessibility data, respectively. For joint data visualization, UMAP was computed using the multi-omic WNN graph (parameter setting: nn.name = “weighted.nn”).
To find markers (genes or peaks) for each identified cell type, we used the function FindAllMarkers (parameter settings: “only.pos = TRUE”). For differentially expressed genes, we used additional parameters (“min.pct = 0.2,” “min.diff.pct = 0.1”) to ensure that the markers are sufficiently expressed in the corresponding cell group. For differentially accessible chromatin regions, we used a logistic regression model and added the total number of fragments as a latent variable, as suggested in the Signac tutorial (by specifying additional parameters: min.pct = 0.05, test.use = “LR,” latent.vars = “atac_peak_region_fragments”). Genes or peaks with adjusted P value (using Bonferroni correction) < 0.05 were retained as the cell type–specific markers.
Variance component modeling of gene expression
We performed variance component analysis to characterize how much gene expression variability could be explained by the patterns of chromatin covariance. We selected genes and peaks that are detected in at least 10% of total pseudobulk samples. Then, for the expression vector of each gene g, we fit the following variance component model as suggested in previous studies (24, 25)
where Yg is the centered and scaled normalized gene expression vector of gene g; P and E are sample-sample correlation matrices computed by chromatin accessibility in promoter (defined as within −1000 to +100 bp from TSSs) and enhancer regions (defined as within ±500 kb around TSSs), respectively; and I, A, and U capture the per-individual, per–age group, and noise terms, respectively. The values of the variance parameters of the model were estimated by the average information restricted likelihood estimation (AIREML; “gaston” R package). To determine the proportion of the variance explained by each variance component, we generated a vector Vg which, by definition, sums to 1
For example, the proportion of the variance in expression for gene g explained by the promoters would be represented by the first element in the Vg vector.
TF motif enrichment analysis
We tested a set of peaks of interest for overrepresentation of each DNA motif from the JASPAR 2020 database (71) by using the FindMotifs function in Signac. The resulting P values are FDR adjusted. The per-cell motif activities were computed by using the Signac wrapper for chromVAR, which identifies motifs associated with variability in chromatin accessibility between cells.
Linking gene-regulatory elements and gene expression
We examined the peak-gene relationships by using pseudobulk samples aggregating ATAC-seq and RNA-seq counts. We created these pseudobulk samples by randomly sampling 500 cells from the entire single-cell dataset as seed cells. For each seed cell, we selected its 49 nearest neighbors within the cells of the same cell type annotations. Therefore, we obtained 500 pseudobulk samples, each of which comprised 50 single cells. The ATAC-seq and RNA-seq counts for pseudobulk samples were obtained by summing peak and gene counts across the respective single-cell members.
We then evaluated the peak-gene linkage scores by using a correlation-based approach (17, 19) applied to these pseudobulk samples. We computed the Spearman correlation between all pairs of peaks and expression of nearby genes (within 0.5 Mbp) by using the LinkPeaks function implemented in Signac (RRID:SCR_021158) and then selected all peak-gene pairs with correlation coefficient |ρ| > 0.3 and FDR-adjusted P < 0.1. In addition, we removed peak-gene links that overlapped the promoter region (defined as within −1000 to +100 bp from the TSS).
To define DORCs (a set of nearby peaks correlated with the expression of a target gene), we ranked genes by the number of significantly associated peaks among the peak-gene associations we identified above and determined five peaks per gene as a cutoff based on the elbow method. To quantify DORC score for a gene, we used the sum of normalized counts in all significantly associated peaks per gene, resulting in a DORC × cell matrix.
Using the pseudobulk samples, we defined a normalized score (from 0 to 1) as the pseudo-age for each cell type based on the proportion of cells found in the six different developmental stages. Taking into consideration the nonlinear developing speed across life span, we used log-scale weights, instead of equal weights, for each developmental stage: log10(1) = 0 for early fetal, log10(3) = 0.48 for late fetal, log10(5) = 0.70 for infancy, log10(7) = 0.85 for childhood, log10(9) = 0.95 for adolescence, and log10(10) = 1 for adulthood.
Pseudotime analysis on neuronal populations
We extracted cells that were annotated as neuronal cell types (including RG, IPC, EN-fetal-early, EN-fetal-late, EN, IN-fetal, IN-MGE, and IN-CGE). We reperformed dimension reduction on the corresponding RNA-seq data and derived UMAP representations based on the top 10 PCs. The rationale behind using only the RNA-seq modality here is that RNA-seq provides higher resolution than ATAC-seq in terms of distinguishing cell types, especially maturely differentiated ones (Fig. 1C), which are usually the end points of trajectories and, hence, are necessary for accurate trajectory inference. In addition, we removed a small population between the terminal clusters of IN-MGE and IN-CGE populations for sharper trajectory reconstruction of neuronal developmental lineages (fig. S3B). Next, we used the UMAP coordinates as input to monocle3 (72) to construct the trajectories across neuronal populations [parameters: learn_graph_control = list(minimal_branch_len = 20), use_partition = FALSE, close_loop = FALSE]. To calculate pseudotime, we selected RGs as the “root” nodes. For cell assignment to lineages, we excluded cells for which the cell type annotations were inconsistent. For example, any inhibitory neurons were excluded from the EN lineage, and any excitatory neurons were excluded from the IN-MGE or IN-CGE lineage. We used the graph_test function in monocle3 to find differentially expressed genes on the trajectory of a specific lineage and selected genes with q value <0.01.
For visualization and residual analysis purposes, we used the DORC scores and the associated gene expression, smoothed over the inferred pseudotime, by applying the tradeSeq package (73) with nine knots. The “knots” are points where a set of basis functions are joined together to create the smoothers. The number of knots was selected to reach an optimal bias-variance trade-off for the smoother, according to the Akaike information criterion (AIC) (fig. S3B). Specifically, we fit a negative binomial generalized additive model (NB-GAM) for every gene in each lineage by using the fitGAM function (parameter setting: knots = 9) and then obtain the estimated smoother by using the predictSmooth function. The residual for each gene was calculated by subtracting the min-max normalized gene expression values from the min-max normalized DORC scores.
Overlap with genetic risk variants
To investigate whether the cell-specific peaks and genes might play a role in various brain and non-brain–related traits, we quantified their colocalization with common risk variants from 53 GWAS (tables S10 and S11). To perform this analysis for snATAC-seq data, we chose the top 2500 most specific peaks per cell type (padded by 1 kb in both directions to include adjacent variants) and applied LD score regression (LD-score) partitioned heritability (41). This method calculates whether common genetic variants located within cell type–specific peaks explain more of the heritability than variants not in those regions when adjusting for the number of variants and also for genetic context (e.g., introns, exons, promoters, or intergenic regions). To perform this analysis for snRNA-seq data, we selected the top 500 most specific genes per each cell type and applied MAGMA (40). Cell-specific genes were padded by 35 kb upstream and 10 kb downstream to also include genetic variants in the proximal regulatory regions. For all GWAS, we excluded the broad major histocompatibility complex region (hg19 coordinates on chr6: 25 to 35 Mb) due to its extensive and complex LD structure. LD was estimated from the European panel of 1000 Genome Project phase 3. For MDD GWAS, we used a version of summary statistics without 23andMe individuals. Summary statistics were downloaded from the designated locations mentioned in the original manuscripts that are referred to in tables S10 and S11.
To decide whether the variants overlapping DORCs and km1 to km4 peaks (i.e., peaks linked to km1 to km4 genes) are associated with significantly higher negative impact in the context of neurodevelopmental disorders, we retrieved 434 disruptive and 366 benign de novo variants prioritized by neural network model BPnet (74) from whole-genome sequencing on autism cases and controls (75). Then, we quantified the significance of the difference between the number of overlaps of disruptive variants and benign variants using the one-sided z test.
To link risk loci associated with neuropsychiatric disorders to their causal genes, we first collected a list of genome-wide significant variants of those disorders extended by the variants that are in strong LD (R2 ≥ 0.8). For this purpose, we used LD matrix for 1000 Genomes Phase 1 of individuals of European ancestry downloaded from https://zenodo.org/record/3404275#.YaQec5HMJdC. Then, we overlapped these variants with the peaks for which we have at least one peak-gene defining the gene under regulation (peaks were lifted to hg19 to match default hg19 coordinates of index SNPs and LD buddies).
Validation experiments
CRISPRi
The CRISPRi algorithm of the Broad Institute Genetic Perturbation Platform was used to design guides against all the targets except the CUX2 Enhancer. The Benchling CRISPR design tool was used to design guides against the CUX2 enhancer (Supplementary Text, “CRISPRi guides”).
Plasmid preparation
The top three guide sequences for every target and their reverse complement sequences were synthesized by IDT (Integrated DNA Technologies) with the appropriate overhang sequences to be cloned downstream of a constitutively expressed U6 promoter in a lentiviral vector (lentiGuide-Hygro-mTagBFP2, Addgene, catalog no. 99374) using the golden gate cloning method. Guide oligos and their reverse complement oligos were initially phosphorylated and annealed using the protocol described in Supplementary Text, “Protocol for plasmid preparation.”
Transduction/differentiation
hiPSC-NPCs were differentiated into forebrain neurons as previously described (36). hiPSC-NPCs were seeded at low density with 1 μl of lentivirus per ml of NPC media [Dulbecco’s modified Eagle’s medium (DMEM)/F12, 1× N2, 1× B27-RA (Invitrogen), laminin (1 μg/ml), and fibroblast growth factor 2 (20 ng/ml) in Matrigel-coated plates]. One to 2 days after plating, cells were treated with hygromycin (1 mg/ml; Thermo Fisher Scientific) to select for cells containing the gRNA. Two days after selection, cells were cultured in neural differentiation medium [DMEM/F12 + GlutaMAX, 1× N2, 1× B27-RA, brain-derived neurotrophic factor (20 ng/ml; PeproTech), glial cell line–derived neurotrophic factor (20 ng/ml; PeproTech), 1 mM dibutyryl-cyclic adenosine monophosphate (Sigma-Aldrich), 200 nM ascorbic acid (Sigma-Aldrich), and laminin (1 μg/ml; Thermo Fisher Scientific)]. NPC-derived hiPSC-(forebrain)-neurons were differentiated for 2 to 6 weeks depending on the assay.
RNAscope setup (plating)
hiPSC-NPCs were plated into eight-well chamber slides (Lab-Tek) at a density of 3.0 × 104 per well for RNAscope. Lentiviral transduction and differentiation was carried out as previously described. Cells were harvested and RNAscope was performed at 2 weeks after differentiation.
RNAscope
Growth medium was removed from the eight-well chamber slides, and cells were washed with 1× PBS (500 μl per chamber). Cells were then fixed with 4% formaldehyde/1× PBS for 30 min at room temperature (500 μl per chamber). After fixation, cells were washed three times with 1× PBS and dehydrated with 50, 70, and then 100% EtOH according to instructions from ACD Biosciences for adherent cells grown in chamber slides. Chambers were removed, and the slides were stored in 100% EtOH at −20°C. Cells were rehydrated, and RNAscope was performed according to the manufacturer’s instructions. Primary probes targeting NEUROD1 [ACD Biosciences RNAscope Probe- Hs-NEUROD1-C2 (no. 437281-C2)] were hybridized and amplified with secondary and tertiary probes, which were then labeled with the fluorophore Opal 570 (Akoya Biosciences FP1488001KT). Primary probes targeting CUX2 (ACD Biosciences RNAscope Probe- Hs-CUX2-C3 (no. 425581-C3) were hybridized and amplified with secondary and tertiary probes, which were then labeled with the fluorophore Opal 690 (Akoya Biosciences FP1497001KT). Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and mounted with ProLong Gold Antifade Reagent (Thermo Fisher Scientific P36934). Mounting medium was allowed to dry overnight, and then slides were stored in the dark at 4°C before cells were imaged.
RNAscope slides were imaged with the Zeiss Axioimager.Z2(M) widefield microscope with a Axiocam monochrome 503 charge-coupled device camera (pixel size: 4.54 μm × 4.54 μm). A 10× air/dry objective [Fluar 10×/numerical aperture (NA) 0.5] was initially used to take snapshots of large fields of cells (400-ms exposure), and then a 63× oil immersion (PlanApo 63×/NA 1.4) objective was used to capture images at higher magnification for further quantitative analysis. With the 63× oil objective, 35 to 40 z stacks (each z stack is 200 nm) were acquired in three channels: Cy5 (Chroma 49006 filter cube) to image CUX2, Cy3 (Chroma 49309 filter cube) to image NEUROD1, and DAPI (Chroma 49000 filter cube). For Cy5 and Cy3, the exposure time for each z stack was 200 ms, and for DAPI, it was 50 ms. Maximum intensity projections were generated from the Cy5 and Cy3 z stacks using FiJi. CUX2 (Cy5) and NEUROD1 (Cy3) RNAscope spots in the maximum intensity projections were localized using the IDL script LocalizeApp, which uses two-dimensional Gaussian fitting to calculate the center of diffraction-limited spots. For the DAPI z stack, the z plane in which nuclei were in focus was used for manual segmentation of nuclei and generation of binary nuclear masks. The IDL script FISHauxiliary was used to quantify the number of diffraction-limited spots in each nuclear mask for both CUX2 and NEUROD1.
Supplementary Material
Acknowledgments
We thank the patients and families who donated material for these studies. Brain tissue for the study was obtained through the NIH Neurobiobank from the following brain bank collections: The Mount Sinai/JJ Peters VA Medical Center NIH Brain and Tissue Repository and the University of Maryland Brain and Tissue Bank. We thank the computational resources and staff expertise provided by the Scientific Computing at the Icahn School of Medicine at Mount Sinai. We thank P. Dong, X. Wang, and other members of the Roussos laboratory for helpful discussion. We thank the computational resources and staff expertise provided by the Scientific Computing of the Icahn School of Medicine at Mount Sinai. This publication was supported by and coordinated through the BICCN. This publication is part of the Human Cell Atlas (www.humancellatlas.org/publications/).
Funding: This work was supported by the National Institute of Mental Health, NIH grants, RF1-MH128970 (to G.-C.Y. and P.R.), R01-MH110921 (to P.R.), U01-MH116442 (to P.R.), R01-MH125246 (to P.R.), and R01-MH109897 (to P.R. and K.J.B.) and the National Institute on Aging, NIH grants R01-AG050986 (to P.R.), R01-AG067025 (to P.R.), and R01-AG065582 (to P.R.). J.B. was supported in part by Alzheimer’s Association Research Fellowship AARF-21-722200.
Author contributions: P.R. conceived and designed the project. J.F.F. and P.R. designed experimental strategies for single-nucleus profiling of human postmortem tissue. N.M.T. performed tissue dissections for fetal specimens. J.F.F. generated single-nucleus multi-ome data. S.R., J.M.V., O.L., A.L., K.J.B., and J.F.F. performed validation experiments. K.Z., J.B., T.C., D.L., G.-C.Y., and P.R. designed analytical strategies. K.Z. conducted initial bioinformatics, sample processing, and quality control for single-nucleus data. K.Z. developed the computational scheme and performed the downstream analysis. J.B., T.C., and C.C. performed the GWAS enrichment analysis and mapping of GWAS loci to causal genes. Data analysis was supervised by D.L., G.-C.Y., and P.R. K.Z., J.B., S.R., G.-C.Y., J.F.F., and P.R. wrote the manuscript with input from all authors.
Competing interests: K.Z. is now an employee of Gilead Sciences. The authors declare that they have no competing interests.
Data and materials availability: Raw data (FASTQ files) and processed data (BigWig files; coordinates of open chromatin regions; count matrices) were deposited in NIH GEO (RRID:SCR_005012) under accession no. GSE204684. UCSC genome browser (RRID:SCR_005780) tracks for cell type pseudobulks are available at http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg38&hubUrl=https://dual-assay.s3.amazonaws.com/hub.txt and from project webpage at https://labs.icahn.mssm.edu/roussos-lab/3dg_dual_assay/. Single-cell data can be further inspected at the Broad Institute Single Cell Portal (RRID:SCR_014816) under accession no SCP1859 (https://singlecell.broadinstitute.org/single_cell/study/SCP1859) and at CELLxGENE (RRID:SCR_021059) portal (https://cellxgene.cziscience.com/collections/ceb895f4-ff9f-403a-b7c3-187a9657ac2c). The code used to perform the analysis described in this study is available at https://doi.org/10.5281/zenodo.7703253. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S4
Legends for tables S1 to S14
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S14
Correction 29 November 2023:
In the original version of the Supplementary Materials, fig. S1 was missing panels (F) and (G). The Supplementary Materials has been updated with the correct figure. The HTML has been updated. The original version is available here:
REFERENCES AND NOTES
- 1.Bae B.-i., Jayaraman D., Walsh C. A., Genetic changes shaping the human brain. Dev. Cell 32, 423–434 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silbereis J. C., Pochareddy S., Zhu Y., Li M., Sestan N., The cellular and molecular landscapes of the developing human central nervous system. Neuron 89, 248–268 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birnbaum R., Weinberger D. R., Genetic insights into the neurodevelopmental origins of schizophrenia. Nat. Rev. Neurosci. 18, 727–740 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Sestan N., State M. W., Lost in translation: Traversing the complex path from genomics to therapeutics in autism spectrum disorder. Neuron 100, 406–423 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang H. J., Kawasawa Y. I., Cheng F., Zhu Y., Xu X., Li M., Sousa A. M. M., Pletikos M., Meyer K. A., Sedmak G., Guennel T., Shin Y., Johnson M. B., Krsnik Ž., Mayer S., Fertuzinhos S., Umlauf S., Lisgo S. N., Vortmeyer A., Weinberger D. R., Mane S., Hyde T. M., Huttner A., Reimers M., Kleinman J. E., Šestan N., Spatio-temporal transcriptome of the human brain. Nature 478, 483–489 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li M., Santpere G., Kawasawa Y. I., Evgrafov O. V., Gulden F. O., Pochareddy S., Sunkin S. M., Li Z., Shin Y., Zhu Y., Sousa A. M. M., Werling D. M., Kitchen R. R., Kang H. J., Pletikos M., Choi J., Muchnik S., Xu X., Wang D., Lorente-Galdos B., Liu S., Giusti-Rodríguez P., Won H., de Leeuw C. A., Pardiñas A. F.; BrainSpan Consortium; PsychENCODE Consortium; PsychENCODE Developmental Subgroup, Hu M., Jin F., Li Y., Owen M. J., O’Donovan M. C., Walters J. T. R., Posthuma D., Reimers M. A., Levitt P., Weinberger D. R., Hyde T. M., Kleinman J. E., Geschwind D. H., Hawrylycz M. J., State M. W., Sanders S. J., Sullivan P. F., Gerstein M. B., Lein E. S., Knowles J. A., Sestan N., Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science 362, eaat7615 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nord A. S., West A. E., Neurobiological functions of transcriptional enhancers. Nat. Neurosci. 23, 5–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan X., Fu Y., Zhou X., Sun L., Yang M., Wang M., Chen R., Wu Q., Yong J., Dong J., Wen L., Qiao J., Wang X., Tang F., Single-cell transcriptome analysis reveals cell lineage specification in temporal-spatial patterns in human cortical development. Sci. Adv. 6, eaaz2978 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eze U. C., Bhaduri A., Haeussler M., Nowakowski T. J., Kriegstein A. R., Single-cell atlas of early human brain development highlights heterogeneity of human neuroepithelial cells and early radial glia. Nat. Neurosci. 24, 584–594 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelley K. W., Pașca S. P., Human brain organogenesis: Toward a cellular understanding of development and disease. Cell 185, 42–61 (2022). [DOI] [PubMed] [Google Scholar]
- 11.Ziats M. N., Rennert O. M., Identification of differentially expressed microRNAs across the developing human brain. Mol. Psychiatry 19, 848–852 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nowakowski T. J., Bhaduri A., Pollen A. A., Alvarado B., Mostajo-Radji M. A., Di Lullo E., Haeussler M., Sandoval-Espinosa C., Liu S. J., Velmeshev D., Ounadjela J. R., Shuga J., Wang X., Lim D. A., West J. A., Leyrat A. A., Kent W. J., Kriegstein A. R., Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanton S., Boyle M. J., He Z., Santel M., Weigert A., Sanchís-Calleja F., Guijarro P., Sidow L., Fleck J. S., Han D., Qian Z., Heide M., Huttner W. B., Khaitovich P., Pääbo S., Treutlein B., Camp J. G., Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature 574, 418–422 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Bhaduri A., Sandoval-Espinosa C., Otero-Garcia M., Oh I., Yin R., Eze U. C., Nowakowski T. J., Kriegstein A. R., An atlas of cortical arealization identifies dynamic molecular signatures. Nature 598, 200–204 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ziffra R. S., Kim C. N., Ross J. M., Wilfert A., Turner T. N., Haeussler M., Casella A. M., Przytycki P. F., Keough K. C., Shin D., Bogdanoff D., Kreimer A., Pollard K. S., Ament S. A., Eichler E. E., Ahituv N., Nowakowski T. J., Single-cell epigenomics reveals mechanisms of human cortical development. Nature 598, 205–213 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herring C. A., Simmons R. K., Freytag S., Poppe D., Moffet J. J. D., Pflueger J., Buckberry S., Vargas-Landin D. B., Clément O., Echeverría E. G., Sutton G. J., Alvarez-Franco A., Hou R., Pflueger C., McDonald K., Polo J. M., Forrest A. R. R., Nowak A. K., Voineagu I., Martelotto L., Lister R., Human prefrontal cortex gene regulatory dynamics from gestation to adulthood at single-cell resolution. Cell 185, 4428–4447.e28 (2022). [DOI] [PubMed] [Google Scholar]
- 17.Ma S., Zhang B., LaFave L. M., Earl A. S., Chiang Z., Hu Y., Ding J., Brack A., Kartha V. K., Tay T., Law T., Lareau C., Hsu Y.-C., Regev A., Buenrostro J. D., Chromatin potential identified by shared single-cell profiling of RNA and chromatin. Cell 183, 1103–1116.e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hao Y., Hao S., Andersen-Nissen E., Mauck W. M. 3rd, Zheng S., Butler A., Lee M. J., Wilk A. J., Darby C., Zager M., Hoffman P., Stoeckius M., Papalexi E., Mimitou E. P., Jain J., Srivastava A., Stuart T., Fleming L. M., Yeung B., Rogers A. J., McElrath J. M., Blish C. A., Gottardo R., Smibert P., Satija R., Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trevino A. E., Sinnott-Armstrong N., Andersen J., Yoon S.-J., Huber N., Pritchard J. K., Chang H. Y., Greenleaf W. J., Pașca S. P., Chromatin accessibility dynamics in a model of human forebrain development. Science 367, eaay1645 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darmanis S., Sloan S. A., Zhang Y., Enge M., Caneda C., Shuer L. M., Hayden Gephart M. G., Barres B. A., Quake S. R., A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. U.S.A. 112, 7285–7290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burke E. E., Chenoweth J. G., Shin J. H., Collado-Torres L., Kim S.-K., Micali N., Wang Y., Colantuoni C., Straub R. E., Hoeppner D. J., Chen H.-Y., Sellers A., Shibbani K., Hamersky G. R., Diaz Bustamante M., Phan B. N., Ulrich W. S., Valencia C., Jaishankar A., Price A. J., Rajpurohit A., Semick S. A., Bürli R. W., Barrow J. C., Hiler D. J., Page S. C., Martinowich K., Hyde T. M., Kleinman J. E., Berman K. F., Apud J. A., Cross A. J., Brandon N. J., Weinberger D. R., Maher B. J., McKay R. D. G., Jaffe A. E., Dissecting transcriptomic signatures of neuronal differentiation and maturation using iPSCs. Nat. Commun. 11, 462 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rakic P., Developmental and evolutionary adaptations of cortical radial glia. Cereb. Cortex 13, 541–549 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Corces M. R., Granja J. M., Shams S., Louie B. H., Seoane J. A., Zhou W., Silva T. C., Groeneveld C., Wong C. K., Cho S. W., Satpathy A. T., Mumbach M. R., Hoadley K. A., Robertson A. G., Sheffield N. C., Felau I., Castro M. A. A., Berman B. P., Staudt L. M., Zenklusen J. C., Laird P. W., Curtis C.; Cancer Genome Atlas Analysis Network, Greenleaf W. J., Chang H. Y., The chromatin accessibility landscape of primary human cancers. Science 362, eaav1898 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bendl J., Hauberg M. E., Girdhar K., Im E., Vicari J. M., Rahman S., Fernando M. B., Townsley K. G., Dong P., Misir R., Kleopoulos S. P., Reach S. M., Apontes P., Zeng B., Zhang W., Voloudakis G., Brennand K. J., Nixon R. A., Haroutunian V., Hoffman G. E., Fullard J. F., Roussos P., The three-dimensional landscape of cortical chromatin accessibility in Alzheimer’s disease. Nat. Neurosci. 25, 1366–1378 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshida H., Lareau C. A., Ramirez R. N., Rose S. A., Maier B., Wroblewska A., Desland F., Chudnovskiy A., Mortha A., Dominguez C., Tellier J., Kim E., Dwyer D., Shinton S., Nabekura T., Qi Y., Yu B., Robinette M., Kim K.-W., Wagers A., Rhoads A., Nutt S. L., Brown B. D., Mostafavi S., Buenrostro J. D., Benoist C.; Immunological Genome Project , Immunological genome project, the cis-regulatory atlas of the mouse immune system. Cell 176, 897–912.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong P., Hoffman G. E., Apontes P., Bendl J., Rahman S., Fernando M. B., Zeng B., Vicari J. M., Zhang W., Girdhar K., Townsley K. G., Misir R.; CommonMind Consortium, Brennand K. J., Haroutunian V., Voloudakis G., Fullard J. F., Roussos P., Population-level variation in enhancer expression identifies disease mechanisms in the human brain. Nat. Genet. 54, 1493–1503 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kosoy R., Fullard J. F., Zeng B., Bendl J., Dong P., Rahman S., Kleopoulos S. P., Shao Z., Girdhar K., Humphrey J., de Paiva Lopes K., Charney A. W., Kopell B. H., Raj T., Bennett D., Kellner C. P., Haroutunian V., Hoffman G. E., Roussos P., Genetics of the human microglia regulome refines Alzheimer’s disease risk loci. Nat. Genet. 54, 1145–1154 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fulco C. P., Nasser J., Jones T. R., Munson G., Bergman D. T., Subramanian V., Grossman S. R., Anyoha R., Doughty B. R., Patwardhan T. A., Nguyen T. H., Kane M., Perez E. M., Durand N. C., Lareau C. A., Stamenova E. K., Aiden E. L., Lander E. S., Engreitz J. M., Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations. Nat. Genet. 51, 1664–1669 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whyte W. A., Orlando D. A., Hnisz D., Abraham B. J., Lin C. Y., Kagey M. H., Rahl P. B., Lee T. I., Young R. A., Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hnisz D., Abraham B. J., Lee T. I., Lau A., Saint-André V., Sigova A. A., Hoke H. A., Young R. A., Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li P., Marshall L., Oh G., Jakubowski J. L., Groot D., He Y., Wang T., Petronis A., Labrie V., Epigenetic dysregulation of enhancers in neurons is associated with Alzheimer’s disease pathology and cognitive symptoms. Nat. Commun. 10, 2246 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veenvliet J. V., Dos Santos M. T. M. A., Kouwenhoven W. M., von Oerthel L., Lim J. L., van der Linden A. J. A., Koerkamp M. J. A. G., Holstege F. C. P., Smidt M. P., Specification of dopaminergic subsets involves interplay of En1 and Pitx3. Development 140, 3373–3384 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Li Y., Wang R., Qiao N., Peng G., Zhang K., Tang K., Han J.-D. J., Jing N., Transcriptome analysis reveals determinant stages controlling human embryonic stem cell commitment to neuronal cells. J. Biol. Chem. 292, 19590–19604 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cubelos B., Sebastián-Serrano A., Beccari L., Calcagnotto M. E., Cisneros E., Kim S., Dopazo A., Alvarez-Dolado M., Redondo J. M., Bovolenta P., Walsh C. A., Nieto M., Cux1 and Cux2 regulate dendritic branching, spine morphology, and synapses of the upper layer neurons of the cortex. Neuron 66, 523–535 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pataskar A., Jung J., Smialowski P., Noack F., Calegari F., Straub T., Tiwari V. K., NeuroD1 reprograms chromatin and transcription factor landscapes to induce the neuronal program. EMBO J. 35, 24–45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flaherty E., Zhu S., Barretto N., Cheng E., Deans P. J. M., Fernando M. B., Schrode N., Francoeur N., Antoine A., Alganem K., Halpern M., Deikus G., Shah H., Fitzgerald M., Ladran I., Gochman P., Rapoport J., Tsankova N. M., McCullumsmith R., Hoffman G. E., Sebra R., Fang G., Brennand K. J., Neuronal impact of patient-specific aberrant NRXN1α splicing. Nat. Genet. 51, 1679–1690 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weiss L. A., Nieto M., The crux of Cux genes in neuronal function and plasticity. Brain Res. 1705, 32–42 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Gray L. T., Yao Z., Nguyen T. N., Kim T. K., Zeng H., Tasic B., Layer-specific chromatin accessibility landscapes reveal regulatory networks in adult mouse visual cortex. eLife 6, e21883 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sullivan P. F., Geschwind D. H., Defining the genetic, genomic, cellular, and diagnostic architectures of psychiatric disorders. Cell 177, 162–183 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Leeuw C. A., Mooij J. M., Heskes T., Posthuma D., MAGMA: Generalized gene-set analysis of GWAS data. PLOS Comput. Biol. 11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bulik-Sullivan B. K., Loh P.-R., Finucane H. K., Ripke S., Yang J., Patterson N., Daly M. J., Price A. L., Neale B. M., LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corces M. R., Shcherbina A., Kundu S., Gloudemans M. J., Frésard L., Granja J. M., Louie B. H., Eulalio T., Shams S., Bagdatli S. T., Mumbach M. R., Liu B., Montine K. S., Greenleaf W. J., Kundaje A., Montgomery S. B., Chang H. Y., Montine T. J., Single-cell epigenomic analyses implicate candidate causal variants at inherited risk loci for Alzheimer’s and Parkinson's diseases. Nat. Genet. 52, 1158–1168 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skene N. G., Bryois J., Bakken T. E., Breen G., Crowley J. J., Gaspar H. A., Giusti-Rodriguez P., Hodge R. D., Miller J. A., Muñoz-Manchado A. B., O’Donovan M. C., Owen M. J., Pardiñas A. F., Ryge J., Walters J. T. R., Linnarsson S., Lein E. S.; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium, Sullivan P. F., Hjerling-Leffler J., Genetic identification of brain cell types underlying schizophrenia. Nat. Genet. 50, 825–833 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh T., Poterba T., Curtis D., Akil H., Al Eissa M., Barchas J. D., Bass N., Bigdeli T. B., Breen G., Bromet E. J., Buckley P. F., Bunney W. E., Bybjerg-Grauholm J., Byerley W. F., Chapman S. B., Chen W. J., Churchhouse C., Craddock N., Cusick C. M., DeLisi L., Dodge S., Escamilla M. A., Eskelinen S., Fanous A. H., Faraone S. V., Fiorentino A., Francioli L., Gabriel S. B., Gage D., Taliun S. A. G., Ganna A., Genovese G., Glahn D. C., Grove J., Hall M.-H., Hämäläinen E., Heyne H. O., Holi M., Hougaard D. M., Howrigan D. P., Huang H., Hwu H.-G., Kahn R. S., Kang H. M., Karczewski K. J., Kirov G., Knowles J. A., Lee F. S., Lehrer D. S., Lescai F., Malaspina D., Marder S. R., McCarroll S. A., McIntosh A. M., Medeiros H., Milani L., Morley C. P., Morris D. W., Mortensen P. B., Myers R. M., Nordentoft M., O’Brien N. L., Olivares A. M., Ongur D., Ouwehand W. H., Palmer D. S., Paunio T., Quested D., Rapaport M. H., Rees E., Rollins B., Satterstrom F. K., Schatzberg A., Scolnick E., Scott L. J., Sharp S. I., Sklar P., Smoller J. W., Sobell J. L., Solomonson M., Stahl E. A., Stevens C. R., Suvisaari J., Tiao G., Watson S. J., Watts N. A., Blackwood D. H., Børglum A. D., Cohen B. M., Corvin A. P., Esko T., Freimer N. B., Glatt S. J., Hultman C. M., McQuillin A., Palotie A., Pato C. N., Pato M. T., Pulver A. E., Clair D. S., Tsuang M. T., Vawter M. P., Walters J. T., Werge T. M., Ophoff R. A., Sullivan P. F., Owen M. J., Boehnke M., O’Donovan M. C., Neale B. M., Daly M. J., Rare coding variants in ten genes confer substantial risk for schizophrenia. Nature 604, 509–516 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu J. M., Satterstrom F. K., Peng M., Brand H., Collins R. L., Dong S., Wamsley B., Klei L., Wang L., Hao S. P., Stevens C. R., Cusick C., Babadi M., Banks E., Collins B., Dodge S., Gabriel S. B., Gauthier L., Lee S. K., Liang L., Ljungdahl A., Mahjani B., Sloofman L., Smirnov A. N., Barbosa M., Betancur C., Brusco A., Chung B. H. Y., Cook E. H., Cuccaro M. L., Domenici E., Ferrero G. B., Gargus J. J., Herman G. E., Hertz-Picciotto I., Maciel P., Manoach D. S., Passos-Bueno M. R., Persico A. M., Renieri A., Sutcliffe J. S., Tassone F., Trabetti E., Campos G., Cardaropoli S., Carli D., Chan M. C. Y., Fallerini C., Giorgio E., Girardi A. C., Hansen-Kiss E., Lee S. L., Lintas C., Ludena Y., Nguyen R., Pavinato L., Pericak-Vance M., Pessah I. N., Schmidt R. J., Smith M., Costa C. I. S., Trajkova S., Wang J. Y. T., Yu M. H. C.; Autism Sequencing Consortium (ASC); Broad Institute Center for Common Disease Genomics (Broad-CCDG); iPSYCH-BROAD Consortium, Cutler D. J., De Rubeis S., Buxbaum J., Daly M. J., Devlin B., Roeder K., Sanders S. J., Talkowski M. E., Rare coding variation provides insight into the genetic architecture and phenotypic context of autism. Nat. Genet. 54, 1320–1331 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leppa V. M., Kravitz S. N., Martin C. L., Andrieux J., Le Caignec C., Martin-Coignard D., DyBuncio C., Sanders S. J., Lowe J. K., Cantor R. M., Geschwind D. H., Rare inherited and de novo CNVs reveal complex contributions to ASD risk in multiplex families. Am. J. Hum. Genet. 99, 540–554 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mojarad B. A., Yin Y., Manshaei R., Backstrom I., Costain G., Heung T., Merico D., Marshall C. R., Bassett A. S., Yuen R. K. C., Genome sequencing broadens the range of contributing variants with clinical implications in schizophrenia. Transl. Psychiatry 11, 84 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gandal M. J., Zhang P., Hadjimichael E., Walker R. L., Chen C., Liu S., Won H., van Bakel H., Varghese M., Wang Y., Shieh A. W., Haney J., Parhami S., Belmont J., Kim M., Losada P. M., Khan Z., Mleczko J., Xia Y., Dai R., Wang D., Yang Y. T., Xu M., Fish K., Hof P. R., Warrell J., Fitzgerald D., White K., Jaffe A. E.; PsychENCODE Consortium, Peters M. A., Gerstein M., Liu C., Iakoucheva L. M., Pinto D., Geschwind D. H., Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 362, eaat8127 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mullins N., Forstner A. J., O’Connell K. S., Coombes B., Coleman J. R. I., Qiao Z., Als T. D., Bigdeli T. B., Børte S., Bryois J., Charney A. W., Drange O. K., Gandal M. J., Hagenaars S. P., Ikeda M., Kamitaki N., Kim M., Krebs K., Panagiotaropoulou G., Schilder B. M., Sloofman L. G., Steinberg S., Trubetskoy V., Winsvold B. S., Won H.-H., Abramova L., Adorjan K., Agerbo E., Al Eissa M., Albani D., Alliey-Rodriguez N., Anjorin A., Antilla V., Antoniou A., Awasthi S., Baek J. H., Bækvad-Hansen M., Bass N., Bauer M., Beins E. C., Bergen S. E., Birner A., Pedersen C. B., Bøen E., Boks M. P., Bosch R., Brum M., Brumpton B. M., Brunkhorst-Kanaan N., Budde M., Bybjerg-Grauholm J., Byerley W., Cairns M., Casas M., Cervantes P., Clarke T.-K., Cruceanu C., Cuellar-Barboza A., Cunningham J., Curtis D., Czerski P. M., Dale A. M., Dalkner N., David F. S., Degenhardt F., Djurovic S., Dobbyn A. L., Douzenis A., Elvsåshagen T., Escott-Price V., Ferrier I. N., Fiorentino A., Foroud T. M., Forty L., Frank J., Frei O., Freimer N. B., Frisén L., Gade K., Garnham J., Gelernter J., Pedersen M. G., Gizer I. R., Gordon S. D., Gordon-Smith K., Greenwood T. A., Grove J., Guzman-Parra J., Ha K., Haraldsson M., Hautzinger M., Heilbronner U., Hellgren D., Herms S., Hoffmann P., Holmans P. A., Huckins L., Jamain S., Johnson J. S., Kalman J. L., Kamatani Y., Kennedy J. L., Kittel-Schneider S., Knowles J. A., Kogevinas M., Koromina M., Kranz T. M., Kranzler H. R., Kubo M., Kupka R., Kushner S. A., Lavebratt C., Lawrence J., Leber M., Lee H.-J., Lee P. H., Levy S. E., Lewis C., Liao C., Lucae S., Lundberg M., MacIntyre D. J., Magnusson S. H., Maier W., Maihofer A., Malaspina D., Maratou E., Martinsson L., Mattheisen M., McCarroll S. A., McGregor N. W., McGuffin P., McKay J. D., Medeiros H., Medland S. E., Millischer V., Montgomery G. W., Moran J. L., Morris D. W., Mühleisen T. W., O’Brien N., O’Donovan C., Loohuis L. M. O., Oruc L., Papiol S., Pardiñas A. F., Perry A., Pfennig A., Porichi E., Potash J. B., Quested D., Raj T., Rapaport M. H., DePaulo J. R., Regeer E. J., Rice J. P., Rivas F., Rivera M., Roth J., Roussos P., Ruderfer D. M., Sánchez-Mora C., Schulte E. C., Senner F., Sharp S., Shilling P. D., Sigurdsson E., Sirignano L., Slaney C., Smeland O. B., Smith D. J., Sobell J. L., Hansen C. S., Artigas M. S., Spijker A. T., Stein D. J., Strauss J. S., Świtkowska B., Terao C., Thorgeirsson T. E., Toma C., Tooney P., Tsermpini E.-E., Vawter M. P., Vedder H., Walters J. T. R., Witt S. H., Xi S., Xu W., Yang J. M. K., Young A. H., Young H., Zandi P. P., Zhou H., Zillich L.; HUNT All-In Psychiatry, Adolfsson R., Agartz I., Alda M., Alfredsson L., Babadjanova G., Backlund L., Baune B. T., Bellivier F., Bengesser S., Berrettini W. H., Blackwood D. H. R., Boehnke M., Børglum A. D., Breen G., Carr V. J., Catts S., Corvin A., Craddock N., Dannlowski U., Dikeos D., Esko T., Etain B., Ferentinos P., Frye M., Fullerton J. M., Gawlik M., Gershon E. S., Goes F. S., Green M. J., Grigoroiu-Serbanescu M., Hauser J., Henskens F., Hillert J., Hong K. S., Hougaard D. M., Hultman C. M., Hveem K., Iwata N., Jablensky A. V., Jones I., Jones L. A., Kahn R. S., Kelsoe J. R., Kirov G., Landén M., Leboyer M., Lewis C. M., Li Q. S., Lissowska J., Lochner C., Loughland C., Martin N. G., Mathews C. A., Mayoral F., McElroy S. L., McIntosh A. M., McMahon F. J., Melle I., Michie P., Milani L., Mitchell P. B., Morken G., Mors O., Mortensen P. B., Mowry B., Müller-Myhsok B., Myers R. M., Neale B. M., Nievergelt C. M., Nordentoft M., Nöthen M. M., O’Donovan M. C., Oedegaard K. J., Olsson T., Owen M. J., Paciga S. A., Pantelis C., Pato C., Pato M. T., Patrinos G. P., Perlis R. H., Posthuma D., Ramos-Quiroga J. A., Reif A., Reininghaus E. Z., Ribasés M., Rietschel M., Ripke S., Rouleau G. A., Saito T., Schall U., Schalling M., Schofield P. R., Schulze T. G., Scott L. J., Scott R. J., Serretti A., Weickert C. S., Smoller J. W., Stefansson H., Stefansson K., Stordal E., Streit F., Sullivan P. F., Turecki G., Vaaler A. E., Vieta E., Vincent J. B., Waldman I. D., Weickert T. W., Werge T., Wray N. R., Zwart J.-A., Biernacka J. M., Nurnberger J. I., Cichon S., Edenberg H. J., Stahl E. A., McQuillin A., Di Florio A., Ophoff R. A., Andreassen O. A., Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat. Genet. 53, 817–829 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kartha V. K., Duarte F. M., Hu Y., Ma S., Chew J. G., Lareau C. A., Earl A., Burkett Z. D., Kohlway A. S., Lebofsky R., Buenrostro J. D., Functional inference of gene regulation using single-cell multi-omics. Cell Genom. 2, 100166 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma N.-X., Puls B., Chen G., Transcriptomic analyses of NeuroD1-mediated astrocyte-to-neuron conversion. Dev. Neurobiol. 82, 375–391 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weiner D. J., Nadig A., Jagadeesh K. A., Dey K. K., Neale B. M., Robinson E. B., Karczewski K. J., O’Connor L. J., Polygenic architecture of rare coding variation across 394,783 exomes. Nature 614, 492–499 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trubetskoy V., Pardiñas A. F., Qi T., Panagiotaropoulou G., Awasthi S., Bigdeli T. B., Bryois J., Chen C.-Y., Dennison C. A., Hall L. S., Lam M., Watanabe K., Frei O., Ge T., Harwood J. C., Koopmans F., Magnusson S., Richards A. L., Sidorenko J., Wu Y., Zeng J., Grove J., Kim M., Li Z., Voloudakis G., Zhang W., Adams M., Agartz I., Atkinson E. G., Agerbo E., Eissa M. A., Albus M., Alexander M., Alizadeh B. Z., Alptekin K., Als T. D., Amin F., Arolt V., Arrojo M., Athanasiu L., Azevedo M. H., Bacanu S. A., Bass N. J., Begemann M., Belliveau R. A., Bene J., Benyamin B., Bergen S. E., Blasi G., Bobes J., Bonassi S., Braun A., Bressan R. A., Bromet E. J., Bruggeman R., Buckley P. F., Buckner R. L., Bybjerg-Grauholm J., Cahn W., Cairns M. J., Calkins M. E., Carr V. J., Castle D., Catts S. V., Chambert K. D., Chan R. C. K., Chaumette B., Cheng W., Cheung E. F. C., Chong S. A., Cohen D., Consoli A., Cordeiro Q., Costas J., Curtis C., Davidson M., Davis K. L., de Haan L., Degenhardt F., De Lisi L. E., Demontis D., Dickerson F., Dikeos D., Dinan T., Djurovic S., Duan J., Ducci G., Dudbridge F., Eriksson J. G., Fañanás L., Faraone S. V., Fiorentino A., Forstner A., Frank J., Freimer N. B., Fromer M., Frustaci A., Gadelha A., Genovese G., Gershon E. S., Giannitelli M., Giegling I., Giusti-Rodríguez P., Godard S., Goldstein J. I., Peñas J. G., González-Pinto A., Gopal S., Gratten J., Green M. F., Greenwood T. A., Guillin O., Gülöksüz S., Gur R. E., Gur R. C., Gutiérrez B., Hahn E., Hakonarson H., Haroutunian V., Hartmann A. M., Harvey C., Hayward C., Henskens F. A., Herms S., Hoffmann P., Howrigan D. P., Ikeda M., Iyegbe C., Joa I., Julià A., Kähler A. K., Kam-Thong T., Kamatani Y., Karachanak-Yankova S., Kebir O., Keller M. C., Kelly B. J., Khrunin A., Kim S.-W., Klovins J., Kondratiev N., Konte B., Kraft J., Kubo M., Kǔinskas V., Kǔinskiene Z. A., Kusumawardhani A., Kuzelova-Ptackova H., Landi S., Lazzeroni L. C., Lee P. H., Legge S. E., Lehrer D. S., Lencer R., Lerer B., Li M., Lieberman J., Light G. A., Limborska S., Liu C.-M., Lönnqvist J., Loughland C. M., Lubinski J., Luykx J. J., Lynham A., Macek Jr M., Mackinnon A., Magnusson P. K. E., Maher B. S., Maier W., Malaspina D., Mallet J., Marder S. R., Marsal S., Martin A. R., Martorell L., Mattheisen M., McCarley R. W., Donald C. M., McGrath J. J., Medeiros H., Meier S., Melegh B., Melle I., Mesholam-Gately R. I., Metspalu A., Michie P. T., Milani L., Milanova V., Mitjans M., Molden E., Molina E., Molto M. D., Mondelli V., Moreno C., Morley C. P., Muntané G., Murphy K. C., Myin-Germeys I., Nenadić I., Nestadt G., Nikitina-Zake L., Noto C., Nuechterlein K. H., O’Brien N. L., O’Neill F. A., Oh S.-Y., Olincy A., Ota V. K., Pantelis C., Papadimitriou G. N., Parellada M., Paunio T., Pellegrino R., Periyasamy S., Perkins D. O., Pfuhlmann B., Pietiläinen O., Pimm J., Porteous D., Powell J., Quattrone D., Quested D., Radant A. D., Rampino A., Rapaport M. H., Rautanen A., Reichenberg A., Roe C., Roffman J. L., Roth J., Rothermundt M., Rutten B. P. F., Saker-Delye S., Salomaa V., Sanjuan J., Santoro M. L., Savitz A., Schall U., Scott R. J., Seidman L. J., Sharp S. I., Shi J., Siever L. J., Sigurdsson E., Sim K., Skarabis N., Slominsky P., So H.-C., Sobell J. L., Söderman E., Stain H. J., Steen N. E., Steixner-Kumar A. A., Stögmann E., Stone W. S., Straub R. E., Streit F., Strengman E., Stroup T. S., Subramaniam M., Sugar C. A., Suvisaari J., Svrakic D. M., Swerdlow N. R., Szatkiewicz J. P., Ta T. M. T., Takahashi A., Terao C., Thibaut F., Toncheva D., Tooney P. A., Torretta S., Tosato S., Tura G. B., Turetsky B. I., Üçok A., Vaaler A., van Amelsvoort T., van Winkel R., Veijola J., Waddington J., Walter H., Waterreus A., Webb B. T., Weiser M., Williams N. M., Witt S. H., Wormley B. K., Wu J. Q., Xu Z., Yolken R., Zai C. C., Zhou W., Zhu F., Zimprich F., Atbaşoğlu E. C., Ayub M., Benner C., Bertolino A., Black D. W., Bray N. J., Breen G., Buccola N. G., Byerley W. F., Chen W. J., Cloninger C. R., Crespo-Facorro B., Donohoe G., Freedman R., Galletly C., Gandal M. J., Gennarelli M., Hougaard D. M., Hwu H.-G., Jablensky A. V., McCarroll S. A., Moran J. L., Mors O., Mortensen P. B., Müller-Myhsok B., Neil A. L., Nordentoft M., Pato M. T., Petryshen T. L., Pirinen M., Pulver A. E., Schulze T. G., Silverman J. M., Smoller J. W., Stahl E. A., Tsuang D. W., Vilella E., Wang S.-H., Xu S.; Indonesia Schizophrenia Consortium; PsychENCODE; Psychosis Endophenotypes International Consortium; SynGO Consortium, Adolfsson R., Arango C., Baune B. T., Belangero S. I., Børglum A. D., Braff D., Bramon E., Buxbaum J. D., Campion D., Cervilla J. A., Cichon S., Collier D. A., Corvin A., Curtis D., Forti M. D., Domenici E., Ehrenreich H., Escott-Price V., Esko T., Fanous A. H., Gareeva A., Gawlik M., Gejman P. V., Gill M., Glatt S. J., Golimbet V., Hong K. S., Hultman C. M., Hyman S. E., Iwata N., Jönsson E. G., Kahn R. S., Kennedy J. L., Khusnutdinova E., Kirov G., Knowles J. A., Krebs M.-O., Laurent-Levinson C., Lee J., Lencz T., Levinson D. F., Li Q. S., Liu J., Malhotra A. K., Malhotra D., Intosh A. M., Quillin A. M., Menezes P. R., Morgan V. A., Morris D. W., Mowry B. J., Murray R. M., Nimgaonkar V., Nöthen M. M., Ophoff R. A., Paciga S. A., Palotie A., Pato C. N., Qin S., Rietschel M., Riley B. P., Rivera M., Rujescu D., Saka M. C., Sanders A. R., Schwab S. G., Serretti A., Sham P. C., Shi Y., Clair D. S., Stefánsson H., Stefansson K., Tsuang M. T., van Os J., Vawter M. P., Weinberger D. R., Werge T., Wildenauer D. B., Yu X., Yue W., Holmans P. A., Pocklington A. J., Roussos P., Vassos E., Verhage M., Visscher P. M., Yang J., Posthuma D., Andreassen O. A., Kendler K. S., Owen M. J., Wray N. R., Daly M. J., Huang H., Neale B. M., Sullivan P. F., Ripke S., Walters J. T. R., O’Donovan M. C.; Schizophrenia Working Group of the Psychiatric Genomics Consortium , Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 604, 502–508 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neuner I., Kupriyanova Y., Stöcker T., Huang R., Posnansky O., Schneider F., Tittgemeyer M., Shah N. J., White-matter abnormalities in Tourette syndrome extend beyond motor pathways. Neuroimage 51, 1184–1193 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Khakh B. S., Astrocyte-neuron interactions in the striatum: Insights on identity, form, and function. Trends Neurosci. 42, 617–630 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jia Y.-F., Wininger K., Peyton L., Ho A. M.-C., Choi D.-S., Astrocytic glutamate transporter 1 (GLT1) deficient mice exhibit repetitive behaviors. Behav. Brain Res. 396, 112906 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao B., Li T., Yang Y., Wang X., Luo T., Shan Y., Zhu Z., Xiong D., Hauberg M. E., Bendl J., Fullard J. F., Roussos P., Li Y., Stein J. L., Zhu H., Common genetic variation influencing human white matter microstructure. Science 372, eabf3736 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Q., Lv H.-W., Yang S., He Y.-Q., Ma Q.-R., Liu J., NEP1-40 alleviates behavioral phenotypes and promote oligodendrocyte progenitor cell differentiation in the hippocampus of cuprizone-induced demyelination mouse model. Neurosci. Lett. 725, 134872 (2020). [DOI] [PubMed] [Google Scholar]
- 59.Robinette T. M., Nicholatos J. W., Francisco A. B., Brooks K. E., Diao R. Y., Sorbi S., Ricca V., Nacmias B., Brieño-Enríquez M. A., Libert S., SIRT1 accelerates the progression of activity-based anorexia. Nat. Commun. 11, 2814 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lombardi L., Blanchet C., Poirier K., Lebrun N., Ramoz N., Rose Moro M., Gorwood P., Bienvenu T., Anorexia nervosa is associated with Neuronatin variants. Psychiatr. Genet. 29, 103–110 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Negraes P. D., Cugola F. R., Herai R. H., Trujillo C. A., Cristino A. S., Chailangkarn T., Muotri A. R., Duvvuri V., Modeling anorexia nervosa: Transcriptional insights from human iPSC-derived neurons. Transl. Psychiatry 7, e1060 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Handel A. E., Chintawar S., Lalic T., Whiteley E., Vowles J., Giustacchini A., Argoud K., Sopp P., Nakanishi M., Bowden R., Cowley S., Newey S., Akerman C., Ponting C. P., Cader M. Z., Assessing similarity to primary tissue and cortical layer identity in induced pluripotent stem cell-derived cortical neurons through single-cell transcriptomics. Hum. Mol. Genet. 25, 989–1000 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stuart T., Srivastava A., Madad S., Lareau C. A., Satija R., Single-cell chromatin state analysis with Signac. Nat. Methods 18, 1333–1341 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., Liu X. S., Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Amemiya H. M., Kundaje A., Boyle A. P., The ENCODE blacklist: Identification of problematic regions of the genome. Sci. Rep. 9, 9354 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wolock S. L., Lopez R., Klein A. M., Scrublet: Computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hafemeister C., Satija R., Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cusanovich D. A., Hill A. J., Aghamirzaie D., Daza R. M., Pliner H. A., Berletch J. B., Filippova G. N., Huang X., Christiansen L., DeWitt W. S., Lee C., Regalado S. G., Read D. F., Steemers F. J., Disteche C. M., Trapnell C., Shendure J., A single-cell atlas of in vivo mammalian chromatin accessibility. Cell 174, 1309–1324.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Waltman L., van Eck N. J., A smart local moving algorithm for large-scale modularity-based community detection. Eur. Phys. J. B 86, 471 (2013). [Google Scholar]
- 70.Becht E., McInnes L., Healy J., Dutertre C.-A., Kwok I. W. H., Ng L. G., Ginhoux F., Newell E. W., Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2018). [DOI] [PubMed] [Google Scholar]
- 71.Fornes O., Castro-Mondragon J. A., Khan A., van der Lee R., Zhang X., Richmond P. A., Modi B. P., Correard S., Gheorghe M., Baranašić D., Santana-Garcia W., Tan G., Chèneby J., Ballester B., Parcy F., Sandelin A., Lenhard B., Wasserman W. W., Mathelier A., JASPAR 2020: Update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48, D87–D92 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D. M., Hill A. J., Zhang F., Mundlos S., Christiansen L., Steemers F. J., Trapnell C., Shendure J., The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Van den Berge K., Roux de Bézieux H., Street K., Saelens W., Cannoodt R., Saeys Y., Dudoit S., Clement L., Trajectory-based differential expression analysis for single-cell sequencing data. Nat. Commun. 11, 1201 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Trevino A. E., Müller F., Andersen J., Sundaram L., Kathiria A., Shcherbina A., Farh K., Chang H. Y., Pașca A. M., Kundaje A., Pașca S. P., Greenleaf W. J., Chromatin and gene-regulatory dynamics of the developing human cerebral cortex at single-cell resolution. Cell 184, 5053–5069.e23 (2021). [DOI] [PubMed] [Google Scholar]
- 75.An J.-Y., Lin K., Zhu L., Werling D. M., Dong S., Brand H., Wang H. Z., Zhao X., Schwartz G. B., Collins R. L., Currall B. B., Dastmalchi C., Dea J., Duhn C., Gilson M. C., Klei L., Liang L., Markenscoff-Papadimitriou E., Pochareddy S., Ahituv N., Buxbaum J. D., Coon H., Daly M. J., Kim Y. S., Marth G. T., Neale B. M., Quinlan A. R., Rubenstein J. L., Sestan N., State M. W., Willsey A. J., Talkowski M. E., Devlin B., Roeder K., Sanders S. J., Genome-wide de novo risk score implicates promoter variation in autism spectrum disorder. Science 362, eaat6576 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S4
Legends for tables S1 to S14
Tables S1 to S14
Correction 29 November 2023:
In the original version of the Supplementary Materials, fig. S1 was missing panels (F) and (G). The Supplementary Materials has been updated with the correct figure. The HTML has been updated. The original version is available here: