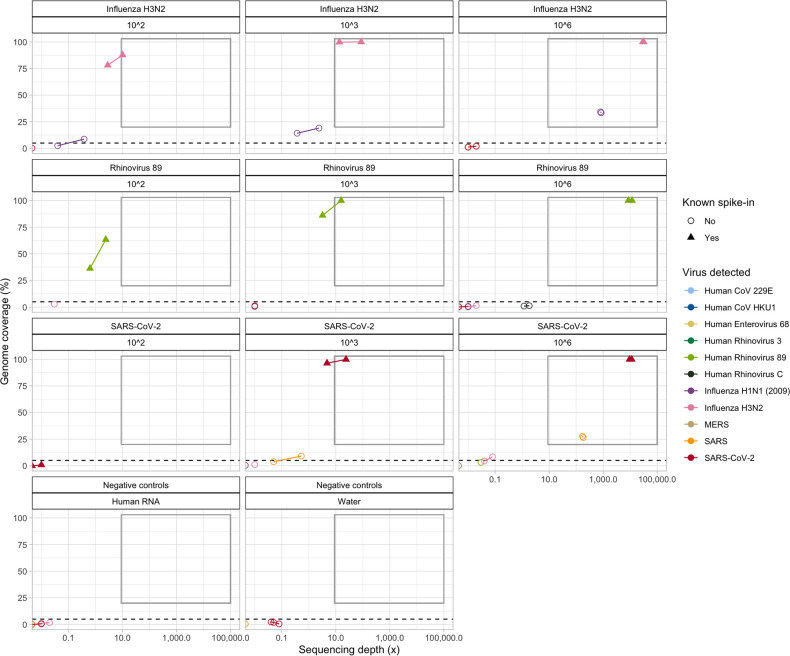
Fig. 1.
OneCodex sequencing depth (×) and genome coverage estimates (%) for each spike-in sample (replicates of influenza H3N2, rhinovirus 89 and SARS-CoV-2 delta), at three different concentrations (102, 103 and 106 viral genome copies µl–1), with eight samples multiplexed. ‘Genome coverage’ denotes the proportion of the relevant viral reference genome covered by at least one sequence; ‘sequencing depth’ denotes the mean sequencing depth across the entire reference genome. Points within the grey box reflect samples which meet the combined OneCodex coverage/depth thresholds used to call a virus ‘detected’. All points above the dashed line but not in the grey box reflect samples that would meet the OneCodex coverage threshold used to call viral presence as ‘indeterminate’. Experiments on the virus dilution series were carried out in duplicate (r1, r2), and on the negative controls in triplicate (water and the universal human RNA used as the background for the spiked samples; r1, r2, r3). Replicate samples (where both samples had evidence of viral sequence detected) are joined by a line. Note the log scale on the x-axis.