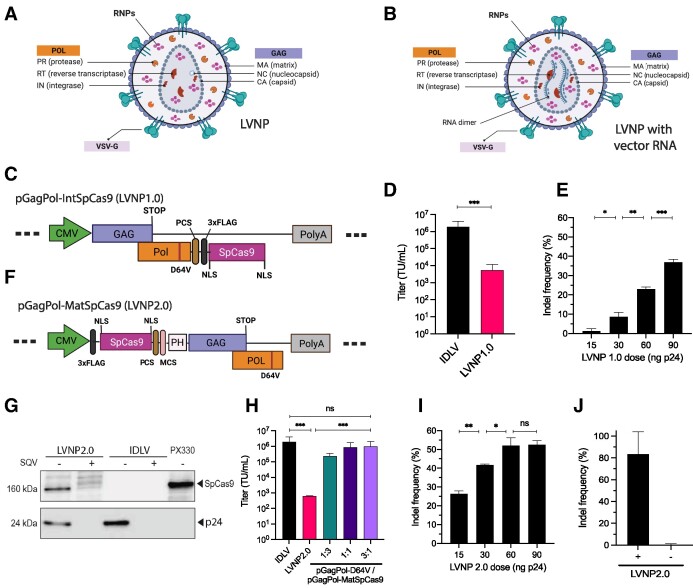
Figure 1.
Incorporation of SpCas9 into lentivirus-derived nanoparticles (LVNPs). (A) Schematic representation of LVNP and (B) LVNP loaded with a vector genome. (C) Schematics of pGagPol-IntSpCas9 composed of FLAG-tagged SpCas9 fused to the C-terminus of the integrase domain (encoded by Pol) and flanked by a PCS for HIV-1 proteolytic release. (D) Functional titers of IDLV/PGK-eGFP and LVNP1.0/PGK-eGFP was evaluated by flow cytometry. (E) Indel frequencies in the AFF1 locus after transduction with increasing dosages of LVNP1.0. (F) Schematics of pGagPol-MatSpCas9 composed of FLAG-tagged SpCas9 fused to the N-terminal of Gag harbouring an intervening phospholipase C-δ1 pleckstrin homology (PH) domain. (G) Western Blot analysis of FLAG-tagged SpCas9 (FLAG antibody) and p24 loading control of purified LVNP2.0 (90 ng p24) and IDLV (90 ng p24) produced in the presence/absence of the HIV-1 protease inhibitor saquinavir (SQV). (H) Estimation of functional titers of LVNP2.0 with/without titration of increasing amounts of pGagPol-D64V. (I) Indel frequencies in the AFF1 locus after transduction with increasing dosages of LVNP2.0 (a 1:3 ratio of pGagPol-D64V:pGagPol-MatSpCas9 was used). (J) Indel frequency after transduction of LVNP2.0 with (+) and without (−) VSV-G pseudotyping. Significant P-values (Mann–Whitney U-test) are marked by *P< 0.05, **P< 0.01, ***P< 0.005. All data is presented as ±SD of triplicates. n.s: non-significant.