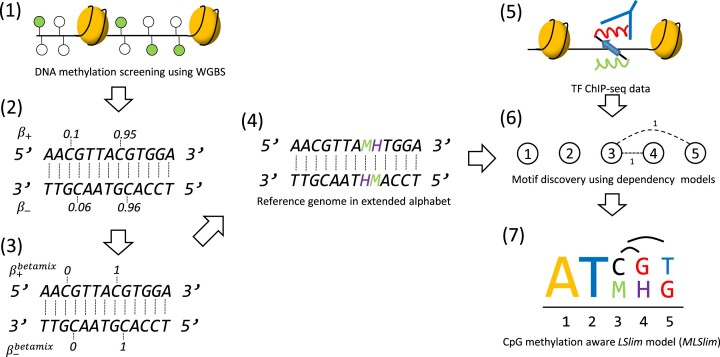
Figure 1.
Overview of the MeDeMo workflow: (1) DNA methylation is assessed using whole genome bisulfite sequencing. (2) DNA methylation is quantified using β-values. (3) Methylation calls (β-values) are discretised using the betamix approach resulting in a binary methylation state for each Cytosine in a CpG context. (4) A novel reference genome is generated by denoting occurrences of methylated cytosines with the letter M and occurrences of guanines opposite of a methylated cytosine with the letter H. (5) In vivo transcription factor binding site information are obtained using peak calls from TF -ChIP-seq data. (6) TF binding data is used for motif discovery with LSlim models on the methylation aware reference genomes; (7) resulting in methylation aware TF motif representations.