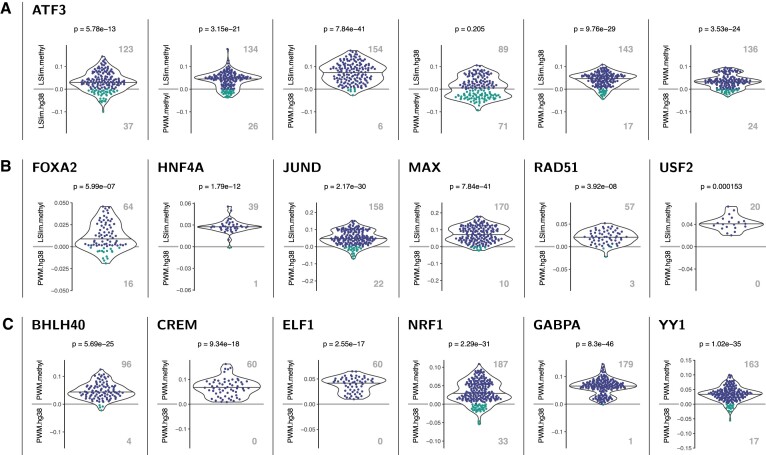
Figure 2.
Examples of TFs with significantly improved classification performance (AUC-PR) in across cell type predictions using a methylation-aware genome. Each panel shows a pairwise comparison of models as indicated by the y-labels above and below the zero line. Each dot represents a case (data sets × cross validation folds) with different colours for positive (i.e., top model performs best) and negative (i.e., bottom model performs best) differences of AUC-PR values. Total number of cases where one model performs better than the other are shown as boldface, grey numbers. In addition, points are summarised by a violin plot and corrected p-values for the H0 that both models perform identical (Prentice test) are given in the header. (A) Pairwise comparison of different modelling variants for ATF3. We find that all methylation-aware models perform better than their counterparts learned on the original hg38 genome and that dependency models (LSlim) perform better than PWM models on the same genome variant. For instance, LSlim.methyl performs better than LSlim.hg38 in 123 cases, whereas the opposite is true for only 37 cases, leading to a P-value of 5.78 × 10−13. (B) Comparison of methylation-aware dependency models (LSlim.methyl) with PWM models using standard hg38 (PWM.hg38) for TFs with a clear advantage of the combination of methylation information and modelling dependencies. (C) Comparison of PWM models learned from the methylation-aware genome with those learned from the standard hg38 genome.