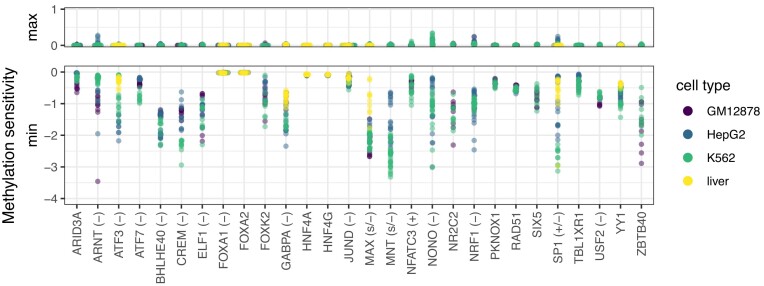
Figure 5.
Methylation within binding motifs is mostly detrimental in models with significantly and consistently improved prediction performance (cf. Figures 2 and 3). For each TF and each data set, we record the profiles of methylation sensitivity as shown in Figure 4. We aggregate this profile to two values per data set by computing the minimum and maximum value of methylation sensitivity, which captures the range of values observed in the profile. Here, we plot these maximum and minimum values of methylation sensitivity across all training data sets. We observe a large amplitude of negative values for the minimum (i.e., methylated DNA being disfavored) but only slightly positive values for the maximum, indicating that—according to the models—DNA methylation is detrimental for the majority of TFs. Methylation sensitivity of TFs according to the literature is given in parentheses (if present), and the corresponding references are given in Table 2.