Figure 7.
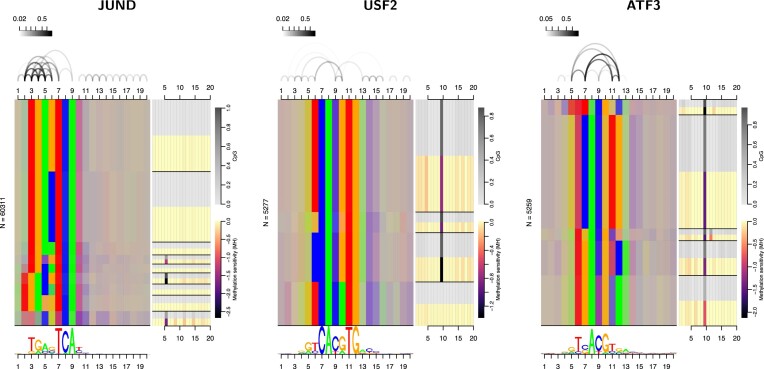
For JUND, USF2 and ATF3, the advantage of combining dependency models with a methylation-aware genome can be attributed to specific properties of the corresponding binding landscapes. For each TF, we visualize predicted binding sites by Dependency Logos that partition binding sites by nucleotides at the most inter-dependent positions and represent each partition using the same colors that are also used in sequence logos (provided below; A=green, C=blue, G=orange, T=red). If a partition contains a mixture of nucleotides at a certain position, colors are mixed as well. For JUND, we find the known variable spacer between the two 3 bp half motif (TGA, TCA), where only the longer spacer frequently contains a CpG. For USF2, the prevalent CpG at positions 9 and 10 shows dependencies to other binding site positions, and is not present in one specific subset (TCACATG) of binding sites. For ATF3, we find broad heterogeneity, where each sub-motif contains CpG at positions 9 and 10 in different proportions.